Team:Valencia Biocampus/modhp
From 2012.igem.org

Modeling for Human Practices: The secret files
Students from our Human Practices subteam found out that two years before releasing their last product, the Talking Life Device, Talking Life Technologies started to carry out a high number of secret experiments regarding the safety of their uprising new sensing-bacteria. These controversial results have just been released two days ago! Click on the image below or in this link to read the complete version of their report!
 |
 |
After analyzing it, we realized that these equations and the parameters used belong to previous studies done in 2010 (1). Since we wanted to test whether these results were true, we have also carried out our own experiments (Table 1) to determine the best data to use and check the possible outcomes. Let’s model Human Practices!
| Table 1. Data from different sources used to modelllize the population dynamics. *Data obtained after several experiments with a cheating plasmid and with no plasmid at all. |
 |
After using our own data the population dynamics we obtained is similar to those from Talking Life Technologies (Figure 4), confirming the worst-case scenario:
 |
| Figure 4. The evolution of both cheater and secretor ("honest") populations using our own data. |
So, how could this big company have done something like that? Is our health under their control?
Experimental evolution prediction from the model
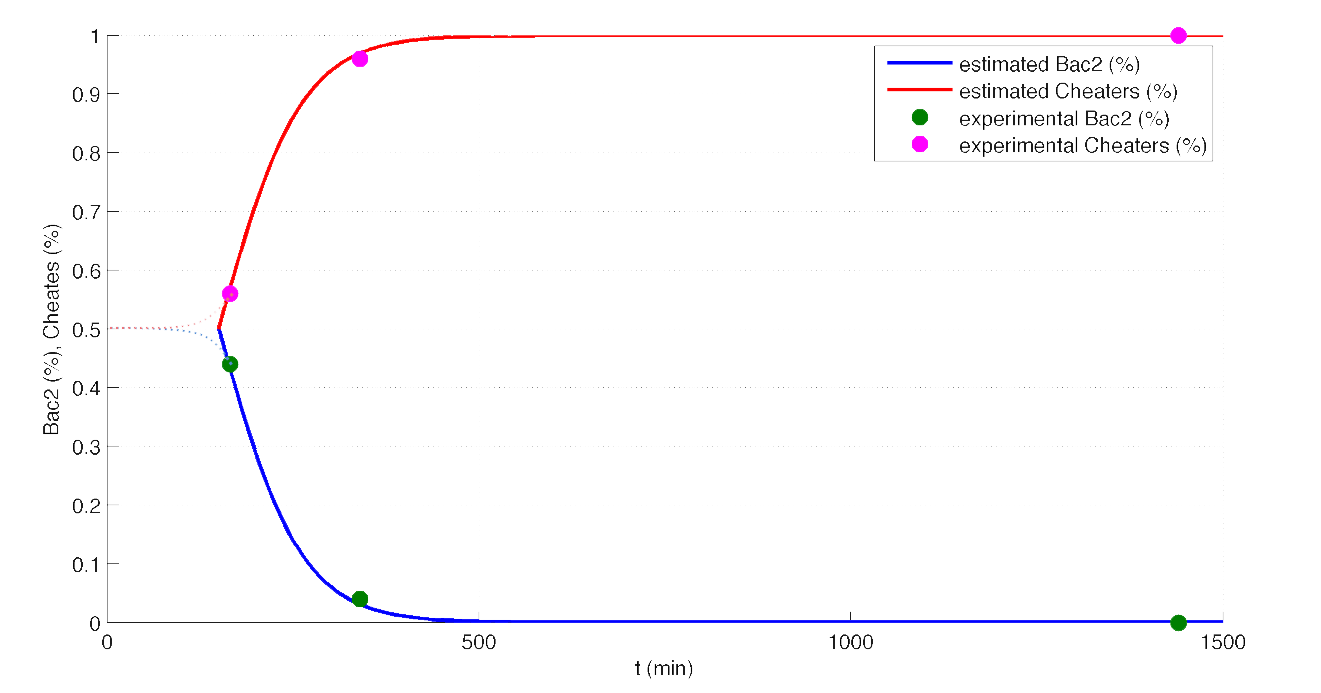
With the model proposed by Frank [1], with some changes to meet our experimental setup, we made a simulation experiment to see what happens when we start with no cheaters. In figure 5, we can see the predicted time evolution of the Bac2 and cheaters population.
| Figure 5. Model prediction of Bac2 and cheaters evolution. |
References:
[1] Frank, A. (2010) Microbial secretor−cheater dynamics. Phil. Trans. R. Soc. B 365, 2515-2522
 "
"