Team:Valencia Biocampus/Bacterium
From 2012.igem.org
(Difference between revisions)
Alejovigno (Talk | contribs) |
Cristina VS (Talk | contribs) (→THE IDEA) |
||
| (132 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
{{:Team:Valencia_Biocampus/banner}} | {{:Team:Valencia_Biocampus/banner}} | ||
| - | {{:Team:Valencia_Biocampus/ | + | {{:Team:Valencia_Biocampus/estiloc3po}} |
{{:Team:Valencia_Biocampus/menu2}} | {{:Team:Valencia_Biocampus/menu2}} | ||
| + | <center> | ||
| + | <ol> | ||
| + | <br> | ||
| + | <div id="Titulos"> | ||
| + | <h2>Bacteria Subteam</h2> | ||
| + | <br> | ||
| + | <div id="PorDefecto"> | ||
| - | == ''' | + | === '''THE IDEA''' === |
| + | <br> | ||
| + | We have developed a simple and fast method of determining "the comfort" of the bacterial culture by asking them four questions related with their metabolic state which are replied in the form of voice answers. Moreover, we have engineered another genetic construction to order them to produce a protein. | ||
| + | Here is an overview of how our bacteria work. For more information look the '''molecular mechanisms''' below. | ||
| - | + | <html> | |
| + | <object classid="clsid:D27CDB6E-AE6D-11cf-96B8-444553540000" codebase="http://download.macromedia.com/pub/shockwave/cabs/flash/swflash.cab#version=4,0,2,0" width="800" height="600" align="middle"> | ||
| + | <param name=wmode value="transparent"> | ||
| + | <param name=movie value="https://static.igem.org/mediawiki/2012/e/e2/Bacteria_Outline_VLC.swf"> | ||
| + | <param name=quality value=high> | ||
| + | <embed src="https://static.igem.org/mediawiki/2012/e/e2/Bacteria_Outline_VLC.swf" wmode=transparent quality=high pluginspage="http://www.macromedia.com/shockwave/download/index.cgi?P1_Prod_Version=ShockwaveFlash" type="application/x-shockwave-flash" width="800" height="600" align="middle"> | ||
| + | </embed> | ||
| + | </object> | ||
| + | </html> | ||
| + | <br><br> | ||
| + | |||
| + | ==='''BACTERIAL SYNTHAXIS'''=== | ||
| + | <br> | ||
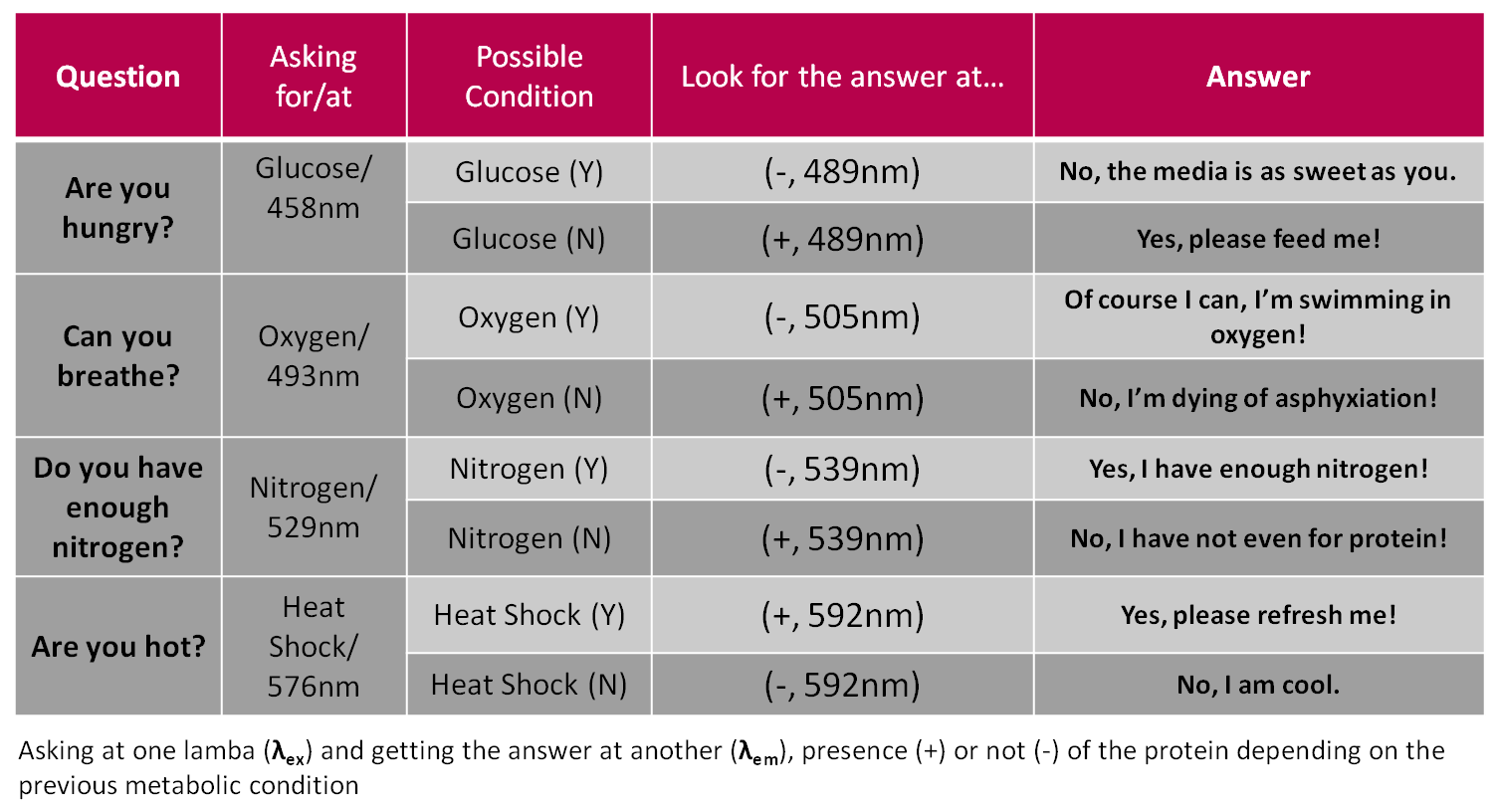
| + | This is the Rosseta stone we used to define both human and bacteria language: | ||
| + | |||
| + | [[Image:Tabla_synthaxis.png|700 px||center]] | ||
| + | <br><br> | ||
| + | |||
| + | ==='''MOLECULAR MECHANISMS'''=== | ||
| + | <br> | ||
| + | Click on each plasmid to learn more about how our constructions work! | ||
| + | <br> | ||
| + | <table align="center" border="0.01" bordercolor="#9F9F9F" style="background-color:#9F9F9F"> | ||
| + | <tr> | ||
| + | <td><html><a href="https://2012.igem.org/Team:Valencia_Biocampus/Molecular#LACTOSE-INDUCED_PROMOTER"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/6/66/Bacteria_hungry.png" width="230" height="200"</a></html></td> | ||
| + | |||
| + | <td><html><a href="https://2012.igem.org/Team:Valencia_Biocampus/Molecular#NITROGEN-REGULATED_PROMOTER"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/76/Bacteria_nitrogen.png" width="230" height="200"</a></html></td> | ||
| + | |||
| + | <td><html><a href="https://2012.igem.org/Team:Valencia_Biocampus/Molecular#OXYGEN-REGULATED_PROMOTER"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/f/f4/Bacteria_oxygen.png" width="230" height="200"</a></html></td> | ||
| + | |||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><html><a href="https://2012.igem.org/Team:Valencia_Biocampus/Molecular#TEMPERATURE-INDUCED_PROMOTER"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/3/38/Bacteria_hot.png" width="230" height="200"</a></html></td> | ||
| + | |||
| + | <td><html><a href="https://2012.igem.org/Team:Valencia_Biocampus/Molecular#UV-INDUCED_PROMOTER"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/3/3d/Bacteria_expressgene.png" width="230" height="200"</a></html></td> | ||
| + | <td><html> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/0/0f/Fondo_gris.png" width="230" height="200"</a></html></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | === '''EXPERIMENTAL OUTLINE''' === | ||
| + | <br> | ||
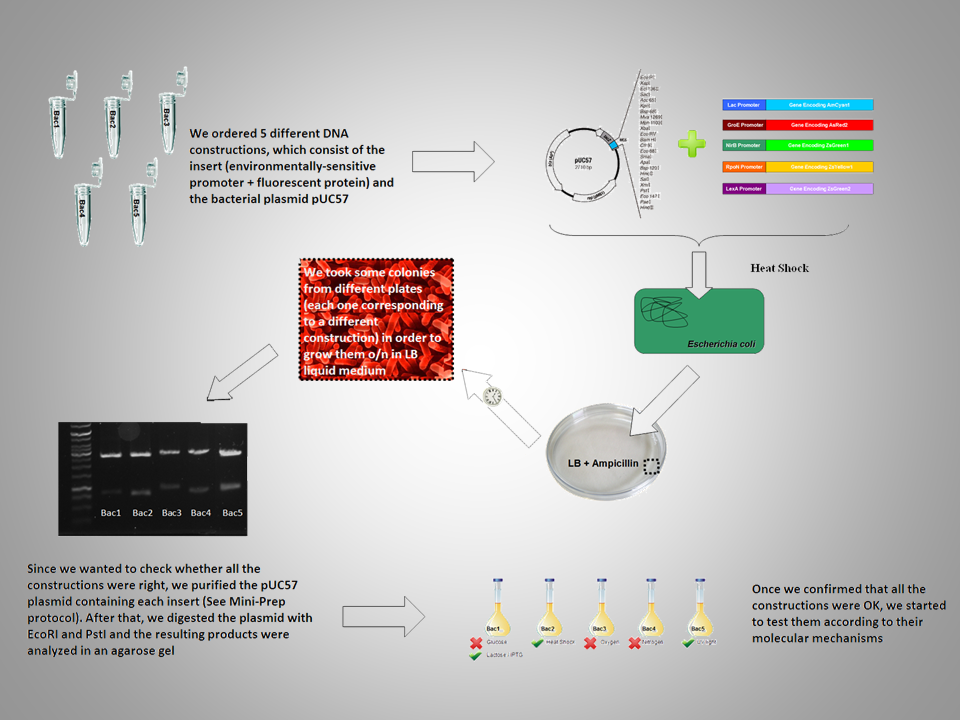
| + | Here you have a graphical overview of the experimental approach we followed: | ||
| + | |||
| + | [[Image:Outline_ppt.png|700 px||center]] | ||
Latest revision as of 19:02, 26 September 2012

Bacteria Subteam
THE IDEA
We have developed a simple and fast method of determining "the comfort" of the bacterial culture by asking them four questions related with their metabolic state which are replied in the form of voice answers. Moreover, we have engineered another genetic construction to order them to produce a protein.
Here is an overview of how our bacteria work. For more information look the molecular mechanisms below.
BACTERIAL SYNTHAXIS
This is the Rosseta stone we used to define both human and bacteria language:
MOLECULAR MECHANISMS
Click on each plasmid to learn more about how our constructions work!
 |
 |
 |
 |
 |
 |
EXPERIMENTAL OUTLINE
Here you have a graphical overview of the experimental approach we followed:
 "
"