|
|
| (109 intermediate revisions not shown) |
| Line 3: |
Line 3: |
| | {{:Team:Valencia_Biocampus/estiloc3po}} | | {{:Team:Valencia_Biocampus/estiloc3po}} |
| | {{:Team:Valencia_Biocampus/menu2}} | | {{:Team:Valencia_Biocampus/menu2}} |
| | + | <center> |
| | | | |
| | + | <ol> |
| | + | <br> |
| | + | <div id="Titulos"> |
| | + | <h2>Bacteria Subteam</h2> |
| | + | <br> |
| | + | <div id="PorDefecto"> |
| | | | |
| - | ==Bacteria Subteam== | + | === '''THE IDEA''' === |
| - | | + | <br> |
| | + | We have developed a simple and fast method of determining "the comfort" of the bacterial culture by asking them four questions related with their metabolic state which are replied in the form of voice answers. Moreover, we have engineered another genetic construction to order them to produce a protein. |
| | Here is an overview of how our bacteria work. For more information look the '''molecular mechanisms''' below. | | Here is an overview of how our bacteria work. For more information look the '''molecular mechanisms''' below. |
| | | | |
| | <html> | | <html> |
| - |
| |
| | <object classid="clsid:D27CDB6E-AE6D-11cf-96B8-444553540000" codebase="http://download.macromedia.com/pub/shockwave/cabs/flash/swflash.cab#version=4,0,2,0" width="800" height="600" align="middle"> | | <object classid="clsid:D27CDB6E-AE6D-11cf-96B8-444553540000" codebase="http://download.macromedia.com/pub/shockwave/cabs/flash/swflash.cab#version=4,0,2,0" width="800" height="600" align="middle"> |
| | + | <param name=wmode value="transparent"> |
| | <param name=movie value="https://static.igem.org/mediawiki/2012/e/e2/Bacteria_Outline_VLC.swf"> | | <param name=movie value="https://static.igem.org/mediawiki/2012/e/e2/Bacteria_Outline_VLC.swf"> |
| | <param name=quality value=high> | | <param name=quality value=high> |
| - | <embed src="https://static.igem.org/mediawiki/2012/e/e2/Bacteria_Outline_VLC.swf" quality=high pluginspage="http://www.macromedia.com/shockwave/download/index.cgi?P1_Prod_Version=ShockwaveFlash" type="application/x-shockwave-flash" width="800" height="600" align="middle"> | + | <embed src="https://static.igem.org/mediawiki/2012/e/e2/Bacteria_Outline_VLC.swf" wmode=transparent quality=high pluginspage="http://www.macromedia.com/shockwave/download/index.cgi?P1_Prod_Version=ShockwaveFlash" type="application/x-shockwave-flash" width="800" height="600" align="middle"> |
| | </embed> | | </embed> |
| | </object> | | </object> |
| - | </center>
| |
| | </html> | | </html> |
| | + | <br><br> |
| | | | |
| | + | ==='''BACTERIAL SYNTHAXIS'''=== |
| | + | <br> |
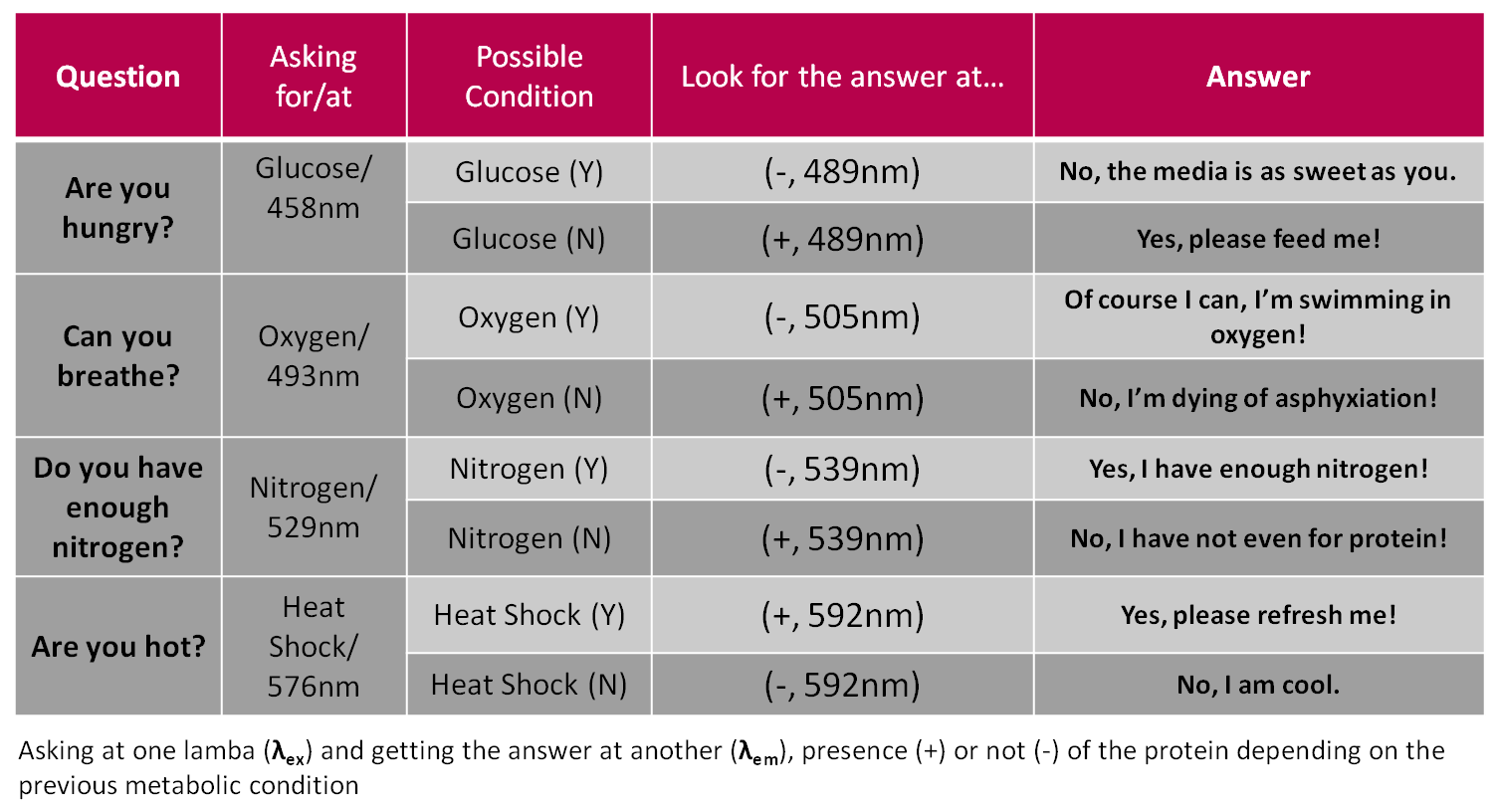
| | + | This is the Rosseta stone we used to define both human and bacteria language: |
| | | | |
| - | ----
| + | [[Image:Tabla_synthaxis.png|700 px||center]] |
| - | | + | <br><br> |
| | | | |
| | ==='''MOLECULAR MECHANISMS'''=== | | ==='''MOLECULAR MECHANISMS'''=== |
| | <br> | | <br> |
| - | '''LACTOSE-INDUCED PROMOTER'''
| + | Click on each plasmid to learn more about how our constructions work! |
| - | <br><br> | + | <br> |
| - | This construction is made up of three parts: (1) the transcription factor-binding site insidethe promoter, (2) the repressor-binding site outside the promoter and (3) the coding sequence, which contains a synthetic fluorescent (blue) protein. Our construction uses the well-known lactose operon system{1}. Since there is an operator region that blocks transcription, it is necessary to know it and avoid it.<br><br>
| + | <table align="center" border="0.01" bordercolor="#9F9F9F" style="background-color:#9F9F9F"> |
| - | When is the protein synthesized? In order to obtain the blue fluorescent protein two conditions have to be met. First condition: there is no glucose in the medium. Second condition: lactose is present in the medium (it also works with other inductors, like IPTG). <br><br>
| + | <tr> |
| - | The molecular mechanism underlying this phenomenon is as follows: a lack of glucose promotes the formation of CAP (or CRP{2}), which binds to specific sites upstream of sugar-metabolizing genes and activates its transcription. The binding of this molecule depends on the presence of the allosteric effector cAMP (the concentration of this metabolite changes in response to the presence or absence of different nutrients).Moreover, another condition is needed since LacI repressor, produced constitutively by the lacI gene inside the lactose operon, will bind to the operator region and will block the transcription unless lactose is also present in the medium.Once lactose enters the cell it is converted to allolactose{3}, and this molecule binds tightly to the repressor so it can no longer block the transcription. Then, the fluorescent protein can glow in the cytoplasm!<br><br>
| + | <td><html><a href="https://2012.igem.org/Team:Valencia_Biocampus/Molecular#LACTOSE-INDUCED_PROMOTER"> |
| - | ¿How did we deal with this construction?In our experiments we not only tested the differential expression when glucose is absent + lactose present and vice versa, but we also tested the expression and growth rates when different compounds were added as carbon-enriched sources. For example, we added sodium acetate and galactose as substitutes of glucose. We determined the best IPTG concentration for our cultures too.
| + | <img src="https://static.igem.org/mediawiki/2012/6/66/Bacteria_hungry.png" width="230" height="200"</a></html></td> |
| - | <br><br> | + | |
| - | References:<br>
| + | <td><html><a href="https://2012.igem.org/Team:Valencia_Biocampus/Molecular#NITROGEN-REGULATED_PROMOTER"> |
| - | 1. F. Jacob and J. Monod. (1959) Genes of structure and genes of regulation in the biosynthesis of proteins. ''C. R. Hebd. Seances Acad''. ''Sci''.249: 1282–4.<br>
| + | <img src="https://static.igem.org/mediawiki/2012/7/76/Bacteria_nitrogen.png" width="230" height="200"</a></html></td> |
| - | 2. S., Busby and R.H., Ebright. (2001). Transcription activation by catabolite activator protein (CAP).'' J. Mol. Biol''.293: 199–213.<br>
| + | |
| - | 3. RE., Huber, K., Wallenfels and G.,Kurz. (1975)The action of beta-galactosidase (Escherichia coli) on allolactose.''Canadian Journal of Biochemistry'', 53(9):1035-1038 <br><br>
| + | <td><html><a href="https://2012.igem.org/Team:Valencia_Biocampus/Molecular#OXYGEN-REGULATED_PROMOTER"> |
| - | https://static.igem.org/mediawiki/2012/d/d6/Tablaglucosa.png <br> | + | <img src="https://static.igem.org/mediawiki/2012/f/f4/Bacteria_oxygen.png" width="230" height="200"</a></html></td> |
| - | ''*For lactose-activation, switch glucose for IPTG.''<br><br>
| + | |
| - | Glucose concentration in lactose-activation media (from a stock of 50 g/L made by Alba & Lamberto): 5 g/L, 1 g/L and 0.1 g/L. Since 0.1 g/L is likely to be the most useful concentration, let’s go on later with 0.05 g/L, 0.01 g/L and 0.001 g/L. Remember to add IPTG!
| + | </tr> |
| - | <br><br> | + | <tr> |
| - | '''NITROGEN-REGULATED PROMOTER''' <br><br>
| + | <td><html><a href="https://2012.igem.org/Team:Valencia_Biocampus/Molecular#TEMPERATURE-INDUCED_PROMOTER"> |
| - | This construction contains two parts: (1) the transcription factor-binding site inside the promoter and (2) the coding sequence, which contains a synthetic fluorescent (yellow) protein. We chose as the promoter sequence the one of the glnA gene. There is long evidence that this promoter is regulated by nitrogen concentration{1}. Moreover, the promoter is not the canonical one (70) but an alternative one (54){2},{3}.<br><br>
| + | <img src="https://static.igem.org/mediawiki/2012/3/38/Bacteria_hot.png" width="230" height="200"</a></html></td> |
| - | When is the protein synthesized? In order to obtain the yellow fluorescent protein a condition should to be met. That condition is related to nitrogen-starvation. The less nitrogen there is, the more expression you get!<br><br>
| + | |
| - | The molecular mechanism underlying this phenomenon is as follows: in gram-negative bacteria, transcriptional activation in response to some external stimuli (absence of nitrogen,high UV-dosis,forexample) often involves the alternative sigma factor, 54. This factor, alsocalledRpoNor Sigma N, was originally identified as the sigma factor for nitrogen-controlled genes. 54works in conjunction with members of the NtrC (Nitrogenregulatoryprotein C) superfamily of transcriptional activators. In our case, when ammonia levels are low the bacterium undergoes some metabolic changes. Within these changes, there are some related to nitrogen assimilation and processing, so our construction, which responds to low ammonia levels, increases its transcription.<br><br>
| + | <td><html><a href="https://2012.igem.org/Team:Valencia_Biocampus/Molecular#UV-INDUCED_PROMOTER"> |
| - | How did we deal with this construction?We developed a nitrogen-gradation tube experiment, which means that we added different (NH4)2SO4 concentrations to each tube, so we can measure a gradation of the protein fluorescence. We used a synthetic medium for this.<br><br>
| + | <img src="https://static.igem.org/mediawiki/2012/3/3d/Bacteria_expressgene.png" width="230" height="200"</a></html></td> |
| | + | <td><html> |
| | + | <img src="https://static.igem.org/mediawiki/2012/0/0f/Fondo_gris.png" width="230" height="200"</a></html></td> |
| | + | </tr> |
| | + | </table> |
| | + | <br> |
| | + | <br> |
| | | | |
| - | References:<br>
| + | === '''EXPERIMENTAL OUTLINE''' === |
| - | 1. Reitzer, L.J., Movsas, B. andMagasanik, B. (1989)Activation of ''glnA'' transcription by Nitrogen Regulator I (NRI)- phosphate in ''Escherichia coli'': evidence for a long-range physical interaction between NRI-phosphate and RNA polymerase.''Journal of bacteriology'', 171 (10):5512-5522<br>
| + | <br> |
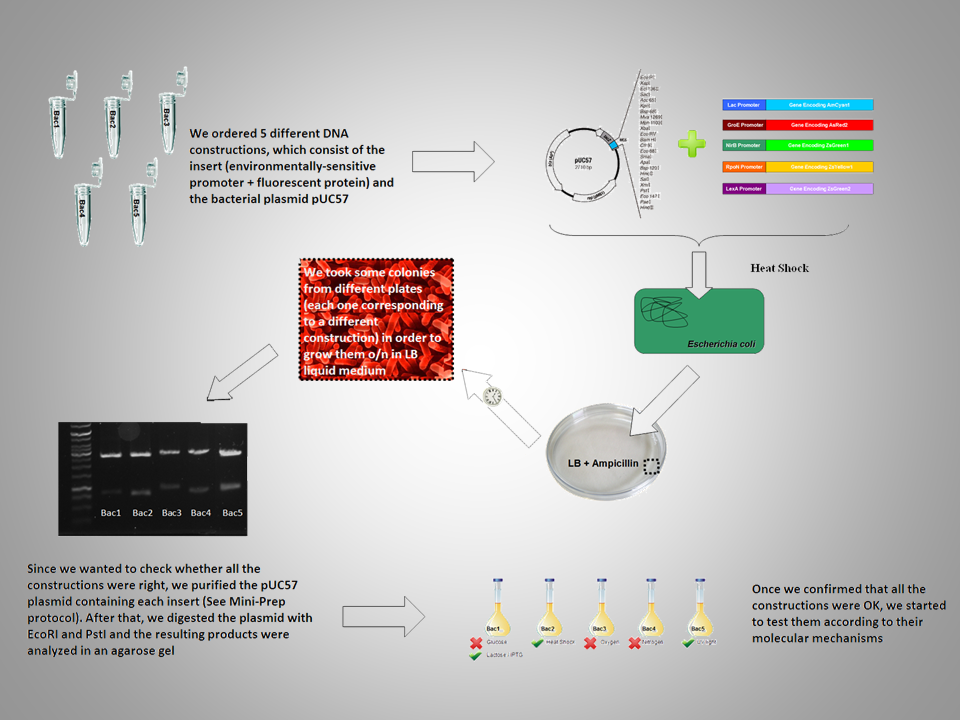
| - | 2. Wang, L., Guo, Y. and Gralla, J.D. (1999) Regulation of sigma 54-dependent transcription by core promoter sequences: role of - 12 region nucleotides. ''Journal of bacteriology'', 181 (24):7558–7565<br>
| + | Here you have a graphical overview of the experimental approach we followed: |
| - | 3. Barrios, H., Valderrama, B. y Morett, E. (1999) Compilation and analysis of 54-dependent promoter sequences. ''Nucleics Acids Research'', 27 (22):4305-4313. <br><br>
| + | |
| - | https://static.igem.org/mediawiki/2012/6/6a/Nitrogen.png <br><br>
| + | |
| - | | + | |
| - | '''OXYGEN-REGULATED PROMOTER''' <br><br>
| + | |
| - | | + | |
| - | This construction has two parts: (1) the transcription factor-binding site inside the promoter and (2) the coding sequence, which contains a synthetic fluorescent (green) protein. We chose as the promoter sequence the one of the nirB gene, because it is 100% dependent on FNR{1},{2}, which is the transcription factor activated when there is no oxygen in the environment. It is involved mainly in cellular respiration and carbon metabolism during anaerobic cell growth.<br><br>
| + | |
| - | When is the protein synthesized? In order to obtain this green fluorescent protein one condition is required: anaerobic conditions should be met. We grew the bacteria in a oxygen-controlled environment{3}.<br><br>
| + | |
| - | The molecular mechanism underlying this phenomenon is as follows: the promoter is activated when there is a lack of O2 in the environment thanks to a family of proteins called FNR. FNR proteins are global transcriptional regulators that respond to fluctuations in ambient oxygen. They recognize DNA sequences in inverted disposition with this sequence: TTGATN1N2N3N4ATCAA{4}, where N1-4 represents a non-conserved tetrad (no influence). The FNR sensor domain contains four cysteine residues that act like ligands for a group (cluster) of 4Fe-4S oxygen-sensitive structures. The acquisition of a 4Fe-4S cluster begins heterodimer formation of FNR that bind to specific sites of DNA and regulate transcription of target promoters. Under aerobic conditions (i.e. when oxygen is present), 4Fe-4S clusters are disassembled and the FNR dimers dissociate and form no structures that can bind to DNA.<br><br>
| + | |
| - | How did we deal with this construction? Anaerobic conditions were achieved after putting a culture of E. coli, stirring, inside a sealed box. We let the bacteria to grow for at least two days and then we measured fluorescence.<br><br>
| + | |
| - | | + | |
| - | References:<br>
| + | |
| | | | |
| - | 1. P.S., Jayaraman, K.L., Gaston, J.A., Cole and S. Busby. (1988) The nirB promoter of ''Escherichia coli'': location of nucleotide sequences essential for regulation by oxygen, the FNR protein and nitrite. ''Mol Microbiol''. 2(4):527-30.<br>
| + | [[Image:Outline_ppt.png|700 px||center]] |
| - | 2. D.F., Browning, J.A. Cole and S. Busby. (2008) Regulation by nucleoid-associated proteins at the ''Escherichia coli'' nir operon promoter. ''J Bacteriol''. 190(21):7258-67.<br>
| + | |
| - | 3. S., Becker, G., Holighaus, T., Gabrielczyk and G., Unden. (1996) O2 as the Regulatory Signal for FNR-Dependent Gene Regulation in'' Escherichia coli. Journal of Bacteriology'', (178): 4515–4521.<br>
| + | |
| - | 4. C., Scott, J.D., Partridge, J.R. Stephenson and J. Green. (2003) DNA target sequence and FNR-dependent gene expression. ''FEBS Letters'' 541: 97-101.<br><br>
| + | |







 "
"