Team:Calgary/Project/HumanPractices/Killswitch/KillGenes
From 2012.igem.org


Hello! iGEM Calgary's wiki functions best with Javascript enabled, especially for mobile devices. We recommend that you enable Javascript on your device for the best wiki-viewing experience. Thanks!
Kill Genes: An active approach

Background
The principal mechanism behind our active killswitch system are exo and endonucleases which work in tandem to cause substantial degradation of the bacterial genome. The chance event of bacterial escape from the bioreactor into tailings ponds triggers the transcription of S7 micrococcal nuclease and CviAII endonuclease.
At the outset of this project, we considered using nucleases which were already present in the registry. Nuclease genes currently in the parts registry proved insufficient for our design parameters for two primary reasons: one, the recognition sites of these enzymes were not frequent enough for widespread genome degradation; and two, the operating temperatures of these enzymes were not optimal for tailings pond conditions. Henceforth, we submitted two new nuclease genes into the registry.
S7 micrococcal nuclease
S7 nuclease is native to Staphylococcus aureus. S. aureus uses this enzyme to destroy extracellular DNA when it infects humans. S7 has both endo and exonuclease activity. This enzyme has a preference for -AT rich regions as opposed to -GC rich regions. However, this enzyme digests the DNA into <200 bp fragments. Ideally this enzyme will be present both intracellularly and extracellularly. We synthesized this enzyme from IDT. However this came with a mutation which altered a lysine residue to an isoleucine thereby making the enzyme dysfunctional.
CviAII restriction enzyme
CviAII is a restriction endonuclease that was sourced from the Chlorella virus PBCV-1 (REF PAPER). Our team selected this enzyme for three reasons.
Firstly, this enzyme recognizes the small four-base restriction site CATG wherein it cuts a staggered end between the A and C on the forward and reverse strands. This is advantageous for the design of our system because of the frequency of this short cut site in the E. coli genome. As opposed to the six base cutter BamHI system submitted by the 2007 Berkely team (BBa_I716462), the CviAII restriction site is to be 16 times more prevalent in the E. coli genome, which translates in finder degradation of the genetic material.
Secondly, the CATG cut site has the probability of being present in start codons of one quarter of genes in the E. coli genome. As such, coding genes will preferentially be selected with activation of CviAII; at this point, the exonuclease activity of S7 micrococcal nuclease can complete degradation of the gene element. Additionally, CviAII is able to cut Dam and Dcm methylated sites in the E. coli genome, and this translates into decreased selectivity of the enzyme.
Finally, the temperature optimum of CviAII functionality is 23 degrees Celsius (REF PAPER). This value is relatively low and better suited to operation the cooler tailings water compared to other systems in the registry. For example, the 2007 Berkely BamHI system is optimized for 37 degrees Celsius and thus would be non-functional the tailings ponds. FIND SOME DATA OF TAILING WATER TEMP.
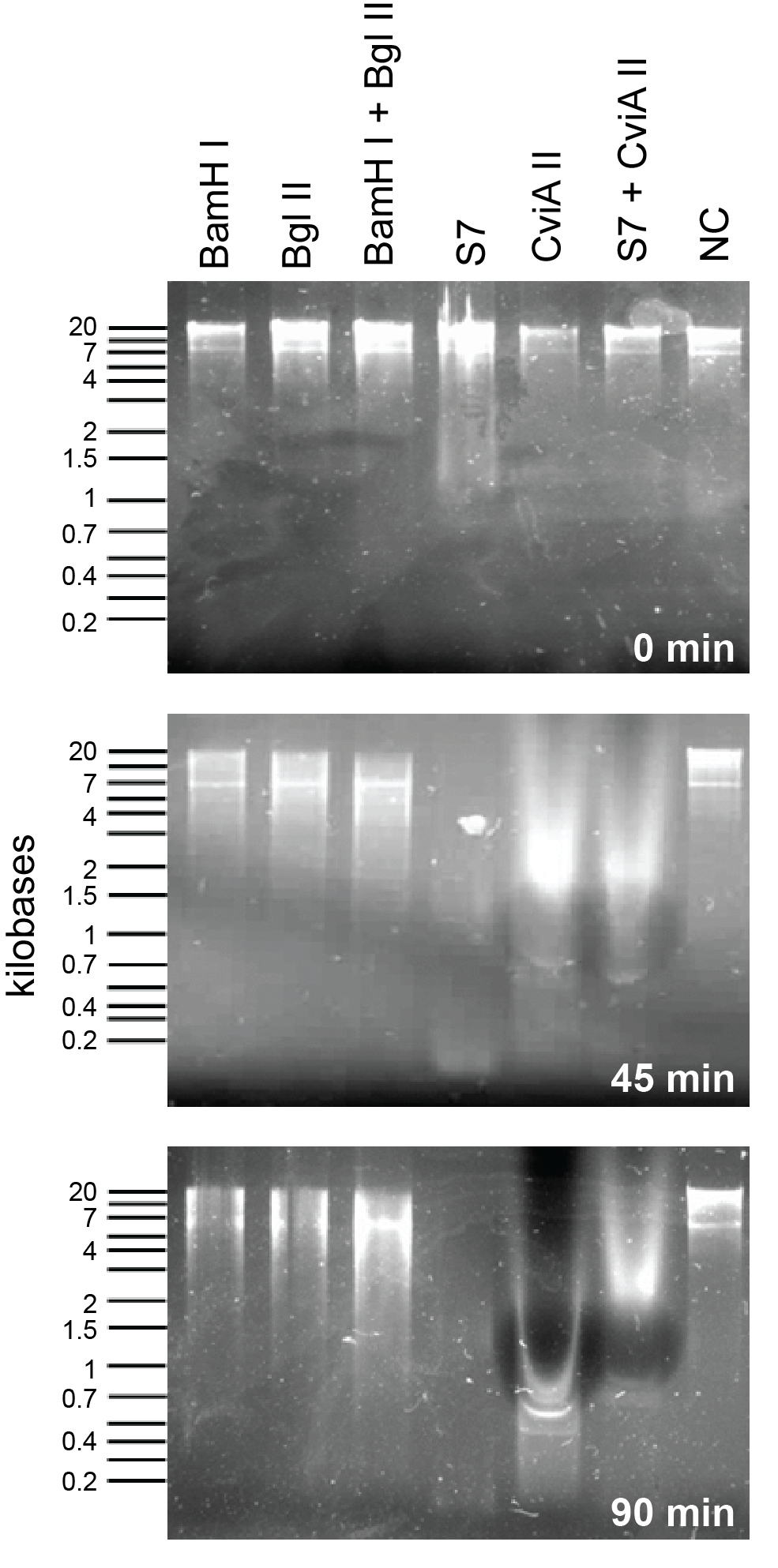
Nuclease assay to evaluate the nucleases present in the registry (BglII and BamHI):
To compare S7 and CviAII to the nucleases already present in the registry we did a nuclease assay with commercially available enzymes from New England Biolabs and an E. coli genomic prep. To see detailed protocol please link see here/link. As can be seen in Figure X, S7 starts acting almost immediately. Within 45 minutes both S7 and CviAII have chewed up the E. coli genome into small fragments whereas BamHI and BglII have sheared the genome into large fragments. Additionally, in 90 minutes, S7 and CviAII have sheared the genome into pieces <200 bp in size whereas there is no difference in the lanes with BglII and BamHI at 90 minutes compared to 45 minutes.
 "
"