Team:Valencia Biocampus/Achievements
From 2012.igem.org
Cristina VS (Talk | contribs) |
|||
| (45 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
{{:Team:Valencia_Biocampus/estiloc3po}} | {{:Team:Valencia_Biocampus/estiloc3po}} | ||
{{:Team:Valencia_Biocampus/menu2}} | {{:Team:Valencia_Biocampus/menu2}} | ||
| - | |||
| - | |||
<center> | <center> | ||
| + | |||
| + | <ol> | ||
<div id="Titulos"> | <div id="Titulos"> | ||
| - | |||
<br> | <br> | ||
| + | <h2>Achievements</h2> | ||
| + | |||
| + | |||
<div id="PorDefecto"> | <div id="PorDefecto"> | ||
| - | + | [[Image:Achievements_vlc.png|700 px||center]] | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
| - | <b> | + | We have successfully talked to microorganisms. Yeast cultures already talk to us when they are hungry (<html><a href="https://2012.igem.org/Team:Valencia_Biocampus/Results2"><b>neat and marked fluorescence</b></a></html> is emitted when there is no glucose in the medium). Taking into account the relevance of yeasts in fermentative processes of the alimentary industry, these results open up new possibilities in the <html><a href="https://2012.igem.org/Team:Valencia_Biocampus/Yeast#INDUSTRIAL_APPLICATIONS"><b>application</b></a></html> of our talking yeasts in the control and monitorization of that kind of processes. Moreover, we achieved true dialogues with <i>E. coli</i>. All our biobricks worked as expected in <i>E. coli</i>, and allowed it to answer the following questions: <html><a href="https://2012.igem.org/Team:Valencia_Biocampus/Results1#Are_you_hot.3F"><b>“Are you hot?”</b></a></html> (thermal shock sensitivity); <html><a href="https://2012.igem.org/Team:Valencia_Biocampus/Results1#Are_you_hungry.3F"><b>“Are you hungry?”</b></a></html> (glucose); <html><a href="https://2012.igem.org/Team:Valencia_Biocampus/Results1#Do_you_have_enough_nitrogen.3F"><b>“Do you have enough nitrogen?”</b></a></html>; and <html><a href="https://2012.igem.org/Team:Valencia_Biocampus/Results1#Can_you_breathe.3F"><b>“Can you breathe?”</b></a></html>. We did so by encoding the questions into light wavelenghths that excite reporter proteins, which were cloned under the control of environment-sensitive promoters. Emitted light pulses were encoded back into the answers “I am cool”, “I am hungry”, etc. We could even make <i>E. coli</i> obey our orders by encoding the <html><a href="https://2012.igem.org/Team:Valencia_Biocampus/Results1#EXPRESS_YOUR_GENE!"><b>“Express your gene”</b></a></html> sentence into a promoter-activation wavelength. |
| - | < | + | |
| - | + | We submitted seven <html><a href="https://2012.igem.org/Team:Valencia_Biocampus/Biobricks">biobricks</a></html> to the registry and fulfilled all other requirements listed at the judging form. Most of our constructions did also work on another bacterial species, <i>Citrobacter freundii</i>: We collaborated with the <html><a href="https://2012.igem.org/Team:Edinburgh" target="blank">Edimburg iGEM team</a></html> sending them our bacterial biobricks (BBa_K763000, BBa_K763001, BBa_K763002, BBa_K763003 & BBa_K763004). They have tested them and the results obtained are collected in <html><a href="https://docs.google.com/document/d/1VykqxS47mSIi9IAc_yiWsosNOt7zBnoxPOEItSdaCTU/edit" target="blank">this</a></html> link. | |
| - | + | ||
| - | </ | + | <br><br> |
| + | |||
| + | == '''Had we to highlight one feature of Talking Life…''' == | ||
| + | |||
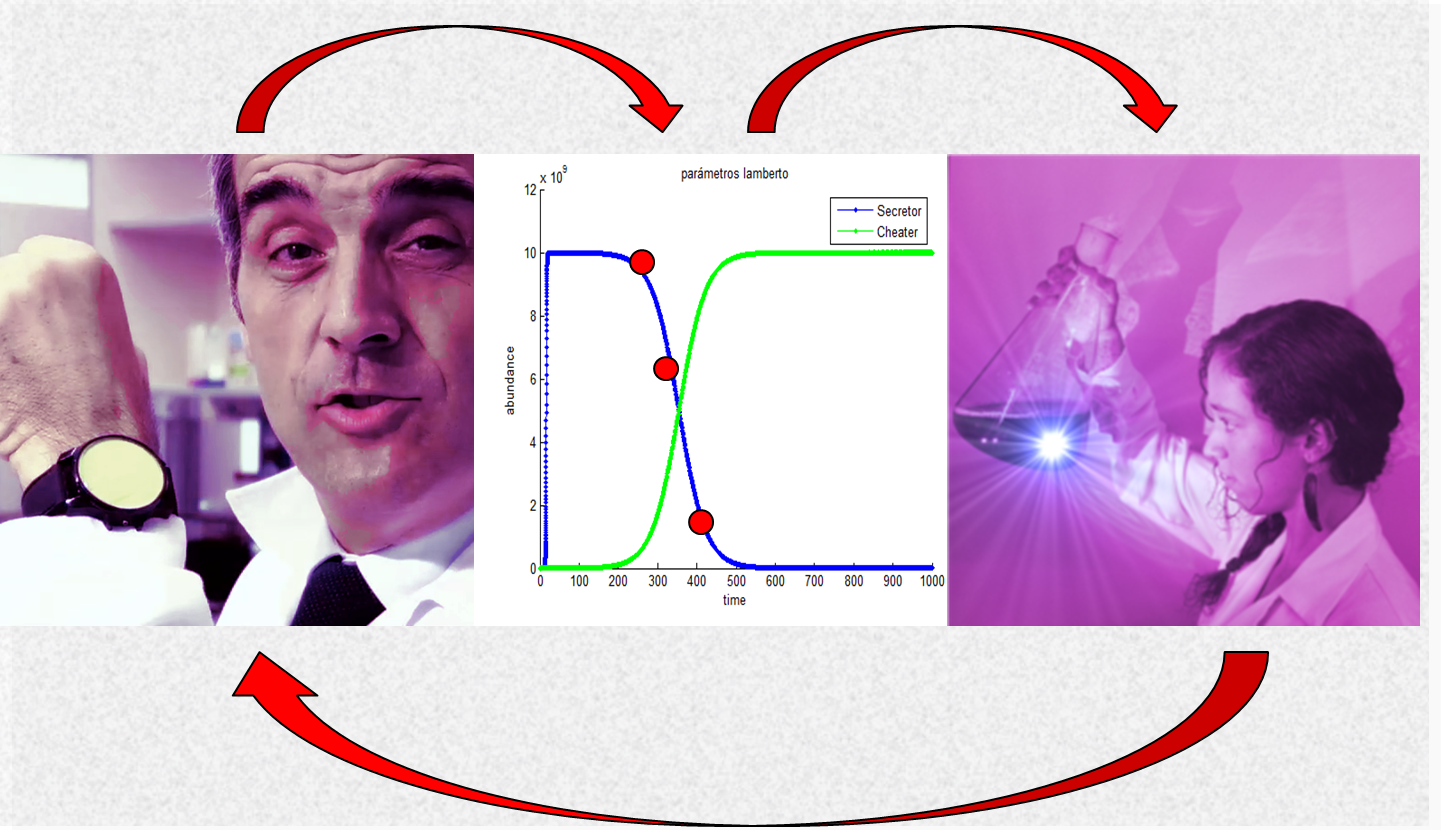
| + | …it would be its integrated nature. In our project, there was a constant flow between dry- and wetlabs, modeling and Human Practices sub-teams. The Human Practices team designed an exercise to identify ethical and social issues and to trigger debate on those issues. We made a short movie. One of the issues that informs the movie is the possibility of cheater mutant bacteria. Complementarily, we carefully modelled the risk of mutant cheaters and | ||
| + | finally end up by discovering that mutant cheaters were in fact among our cultures! We characterized such mutants, performed experimental evolution tests and found them to behave as expected/modeled/feared: mutants actually display an evolutionary advantage on “honest”, heterologous protein-expressing clones. The consequences of all this were already anticipated in the script of our movie, one of the first things we attempted in this challenging project, and maybe the best way to close it: | ||
| + | |||
| + | |||
| + | |||
| + | <html> | ||
| + | <center> | ||
| + | <iframe width="640" height="360" src="http://www.youtube.com/embed/PW4o7-V7KeY" frameborder="0" allowfullscreen></iframe> | ||
| + | </center> | ||
| + | </html> | ||
<br> | <br> | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
Latest revision as of 23:40, 26 September 2012

Achievements
We have successfully talked to microorganisms. Yeast cultures already talk to us when they are hungry (neat and marked fluorescence is emitted when there is no glucose in the medium). Taking into account the relevance of yeasts in fermentative processes of the alimentary industry, these results open up new possibilities in the application of our talking yeasts in the control and monitorization of that kind of processes. Moreover, we achieved true dialogues with E. coli. All our biobricks worked as expected in E. coli, and allowed it to answer the following questions: “Are you hot?” (thermal shock sensitivity); “Are you hungry?” (glucose); “Do you have enough nitrogen?”; and “Can you breathe?”. We did so by encoding the questions into light wavelenghths that excite reporter proteins, which were cloned under the control of environment-sensitive promoters. Emitted light pulses were encoded back into the answers “I am cool”, “I am hungry”, etc. We could even make E. coli obey our orders by encoding the “Express your gene” sentence into a promoter-activation wavelength.
We submitted seven biobricks to the registry and fulfilled all other requirements listed at the judging form. Most of our constructions did also work on another bacterial species, Citrobacter freundii: We collaborated with the Edimburg iGEM team sending them our bacterial biobricks (BBa_K763000, BBa_K763001, BBa_K763002, BBa_K763003 & BBa_K763004). They have tested them and the results obtained are collected in this link.
Had we to highlight one feature of Talking Life…
…it would be its integrated nature. In our project, there was a constant flow between dry- and wetlabs, modeling and Human Practices sub-teams. The Human Practices team designed an exercise to identify ethical and social issues and to trigger debate on those issues. We made a short movie. One of the issues that informs the movie is the possibility of cheater mutant bacteria. Complementarily, we carefully modelled the risk of mutant cheaters and finally end up by discovering that mutant cheaters were in fact among our cultures! We characterized such mutants, performed experimental evolution tests and found them to behave as expected/modeled/feared: mutants actually display an evolutionary advantage on “honest”, heterologous protein-expressing clones. The consequences of all this were already anticipated in the script of our movie, one of the first things we attempted in this challenging project, and maybe the best way to close it:
 "
"