Team:Calgary/Project/OSCAR/Desulfurization
From 2012.igem.org
| Line 9: | Line 9: | ||
<p> | <p> | ||
Sulfur is the third most abundant element in crude oil (Ma, 2010). When sulfur containing hydrocarbons in fuel are burned, S02 and S03 are released into the atmosphere, and are major component in acid rain and dry acid deposition (Reichmuth, 2000). Acid rain (rain with a pH less than 5.3) can cause acidification of aquatic and terrestrial environments, causing nutrient leeching and damaging the health and habitats of many species not tolerant to acidification. | Sulfur is the third most abundant element in crude oil (Ma, 2010). When sulfur containing hydrocarbons in fuel are burned, S02 and S03 are released into the atmosphere, and are major component in acid rain and dry acid deposition (Reichmuth, 2000). Acid rain (rain with a pH less than 5.3) can cause acidification of aquatic and terrestrial environments, causing nutrient leeching and damaging the health and habitats of many species not tolerant to acidification. | ||
| - | Stringent regulations on sulfur content in fossil fuels have been put into place. As of 2005, low-sulfur gasoline (gasoline with a sulfur content of less than 30 mg/kg) is required across all of Canada (Source: Environment Canada). The current process of hydrodesulfurization is environmentally unsound (requiring high temperature and pressure) as well as being quite costly | + | Stringent regulations on sulfur content in fossil fuels have been put into place. As of 2005, low-sulfur gasoline (gasoline with a sulfur content of less than 30 mg/kg) is required across all of Canada (Source: Environment Canada). Naphthenic acid compounds found in the tailings ponds can also be sulfur-containing heterocycles. Because our aim is to convert these compounds to useable fuel, these sulfur atoms must therefore be removed from the naphthenic acids before this goal can be realized. In addition, removal of the sulfur atom in a heterocycle alters the connectivity of the carbon skeleton of the molecule, changing it from a 3-ring compound to a 2-ring compound. |
| - | + | ||
| - | In contrast to | + | <h3>Our Vision</h3><p> |
| - | The 4S pathway found in Rhodococcus spp. has been characterized and shown to remove sulfur from the model substrate dibenzothiophene (DBT) and convert it to 2-hydroxybiphenyl. | + | The current process of hydrodesulfurization used to remove sulfur during fuel upgrading is environmentally unsound (requiring high temperature and pressure) as well as being quite costly <b>Reference, actual figure</b>. In contrast to this, microbes have been found capable of harvesting the sulfur out of hydrocarbons, a process which on an industrial scale would be much more environmentally sound and most likely be less costly to carry out. Though a few pathways for biodesulfurization exist in the microbial world, most involve the destruction of part of the carbon skeleton (an example would be the Kodama pathway). This is unwanted in our system, because by removing some of the carbon backbone to be turned into biomass, the cells effectively reduce the fuel quality.</p> |
| + | <p>The pathway we have chosen for our system is not destructive in this fashion, and therefore preserves (and improves) fuel quality. The 4S pathway found in Rhodococcus spp. has been characterized and shown to remove sulfur from the model substrate dibenzothiophene (DBT) and convert it to 2-hydroxybiphenyl in a non-destructive manner. The pathway has also been shown to act upon derivatives of DBT, and thus we hope that it will also be capable of acting upon naphthenic acids containing sulfur atoms in their structure. | ||
</p><p> | </p><p> | ||
| Line 19: | Line 20: | ||
<h3>4S pathway</h3> | <h3>4S pathway</h3> | ||
<p> | <p> | ||
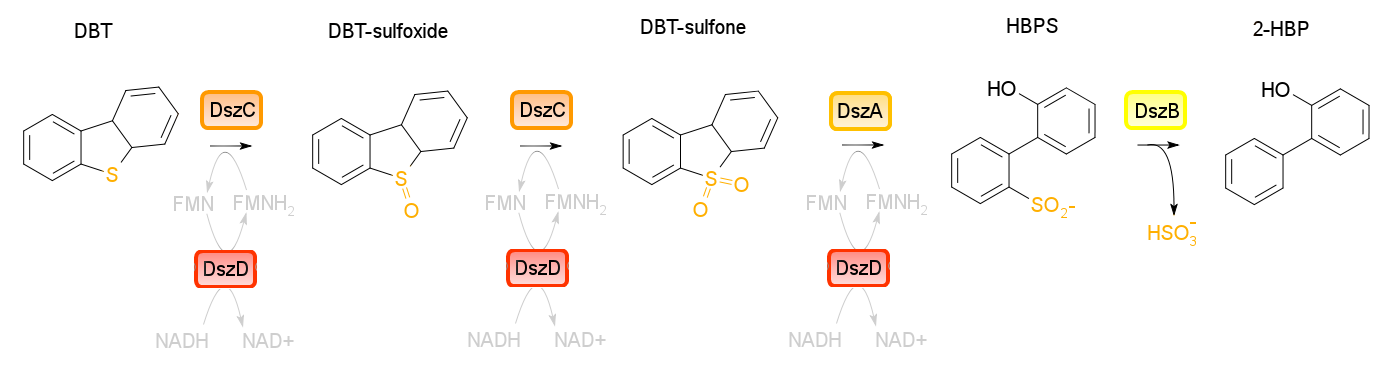
| - | Four enzymes are involved in the 4S pathway. | + | Four enzymes are involved in the 4S pathway, 3 of which are directly involved in the conversion of DBT to 2-HBP. Dibenzothiophene monooxygenase (DszC) is responsible for the first two steps of the pathway, converting DBT to DBT-sulfoxide and finally to DBT-sulfone (DBTO2) through the addition of oxygens to the sulfur atom. DBT-sulfone monooxygenase (DszA) then carries out the next step in the pathway, producing 2-hydroxybiphenyl-2-sulfinic acid (HBPS) through addition of a final oxygen to the heteroatom. This causes cleavage of the chemical bonds at the heteroatom, breaking the ring and converting the compound from a 3-ring structure to a 2-ring structure. HBPS is then converted to the final product of the 4S pathway by HBPS desulfinase (DszB), producing 2-hydroxybiphenyl. At this point, the sulfur has been released from the hydrocarbon in the form of sulfite.</p><p> |
| + | The first three steps of the 4S pathway carried out by the monooxygenase enzymes require FMNH2, and therefore use up the reductive power of the cell. Without a way to recover this, the pathway essentially chokes, desulfurization rates are very low, and cell health suffers. In order to fix this problem, an oxidoreductase (DszD) uses NADH to recycle the FMNH2, allowing the reaction to proceed.</p><p> | ||
| + | <i>dszA</i>, <i>dszB</i> and <i>dszC</i> form an operon on the pSOX plasmid of <i>R. erythropolis</i> while <i>dszD</i> (a flavin oxidoreductase) is present on the chromosomal DNA of the organism. In natural circumstances the 4S pathway has been found to be quite slow, preventing this method of biodesulfurization from becoming commercially feasable. This is due to a number of limitations found in the native setup of the system. With synthetic biology approaches however, it is possible to optimize this pathway by making some alterations. </p> | ||
<h3>Our Approach</h3> | <h3>Our Approach</h3> | ||
| - | <h4>Find an organism</h4> | + | <h4>1) Find an organism</h4> |
<p>Plasmid was isolated(miniprepped) from and Rhodococcus. (I am not sure what is the name of the strain we got from Dr. Gieg). Using primers designed for amplification of each of dszA, dszB, dszC and hpaC these genes were isolated. The primers were designed in a way to add biobrick cut sites to genes. Afterwards, each gene was biobricked into a psb1c3 backbone. The next step was to construct the genes into an operon. However, before construction we had to mutate the enzymes. The promoter controlling the transcription of this pathway is repressed by sulphur containing compounds such as cysteine, methionine, and sulphate (Li et al. 1996). This problem is easily solved since our final construct will have a different promoter from the registry.</p> | <p>Plasmid was isolated(miniprepped) from and Rhodococcus. (I am not sure what is the name of the strain we got from Dr. Gieg). Using primers designed for amplification of each of dszA, dszB, dszC and hpaC these genes were isolated. The primers were designed in a way to add biobrick cut sites to genes. Afterwards, each gene was biobricked into a psb1c3 backbone. The next step was to construct the genes into an operon. However, before construction we had to mutate the enzymes. The promoter controlling the transcription of this pathway is repressed by sulphur containing compounds such as cysteine, methionine, and sulphate (Li et al. 1996). This problem is easily solved since our final construct will have a different promoter from the registry.</p> | ||
Revision as of 00:36, 19 September 2012


Hello! iGEM Calgary's wiki functions best with Javascript enabled, especially for mobile devices. We recommend that you enable Javascript on your device for the best wiki-viewing experience. Thanks!
Desulfurization
Why Remove Sulfur?
Sulfur is the third most abundant element in crude oil (Ma, 2010). When sulfur containing hydrocarbons in fuel are burned, S02 and S03 are released into the atmosphere, and are major component in acid rain and dry acid deposition (Reichmuth, 2000). Acid rain (rain with a pH less than 5.3) can cause acidification of aquatic and terrestrial environments, causing nutrient leeching and damaging the health and habitats of many species not tolerant to acidification. Stringent regulations on sulfur content in fossil fuels have been put into place. As of 2005, low-sulfur gasoline (gasoline with a sulfur content of less than 30 mg/kg) is required across all of Canada (Source: Environment Canada). Naphthenic acid compounds found in the tailings ponds can also be sulfur-containing heterocycles. Because our aim is to convert these compounds to useable fuel, these sulfur atoms must therefore be removed from the naphthenic acids before this goal can be realized. In addition, removal of the sulfur atom in a heterocycle alters the connectivity of the carbon skeleton of the molecule, changing it from a 3-ring compound to a 2-ring compound.
Our Vision
The current process of hydrodesulfurization used to remove sulfur during fuel upgrading is environmentally unsound (requiring high temperature and pressure) as well as being quite costly Reference, actual figure. In contrast to this, microbes have been found capable of harvesting the sulfur out of hydrocarbons, a process which on an industrial scale would be much more environmentally sound and most likely be less costly to carry out. Though a few pathways for biodesulfurization exist in the microbial world, most involve the destruction of part of the carbon skeleton (an example would be the Kodama pathway). This is unwanted in our system, because by removing some of the carbon backbone to be turned into biomass, the cells effectively reduce the fuel quality.
The pathway we have chosen for our system is not destructive in this fashion, and therefore preserves (and improves) fuel quality. The 4S pathway found in Rhodococcus spp. has been characterized and shown to remove sulfur from the model substrate dibenzothiophene (DBT) and convert it to 2-hydroxybiphenyl in a non-destructive manner. The pathway has also been shown to act upon derivatives of DBT, and thus we hope that it will also be capable of acting upon naphthenic acids containing sulfur atoms in their structure.
4S pathway
Four enzymes are involved in the 4S pathway, 3 of which are directly involved in the conversion of DBT to 2-HBP. Dibenzothiophene monooxygenase (DszC) is responsible for the first two steps of the pathway, converting DBT to DBT-sulfoxide and finally to DBT-sulfone (DBTO2) through the addition of oxygens to the sulfur atom. DBT-sulfone monooxygenase (DszA) then carries out the next step in the pathway, producing 2-hydroxybiphenyl-2-sulfinic acid (HBPS) through addition of a final oxygen to the heteroatom. This causes cleavage of the chemical bonds at the heteroatom, breaking the ring and converting the compound from a 3-ring structure to a 2-ring structure. HBPS is then converted to the final product of the 4S pathway by HBPS desulfinase (DszB), producing 2-hydroxybiphenyl. At this point, the sulfur has been released from the hydrocarbon in the form of sulfite.
The first three steps of the 4S pathway carried out by the monooxygenase enzymes require FMNH2, and therefore use up the reductive power of the cell. Without a way to recover this, the pathway essentially chokes, desulfurization rates are very low, and cell health suffers. In order to fix this problem, an oxidoreductase (DszD) uses NADH to recycle the FMNH2, allowing the reaction to proceed.
dszA, dszB and dszC form an operon on the pSOX plasmid of R. erythropolis while dszD (a flavin oxidoreductase) is present on the chromosomal DNA of the organism. In natural circumstances the 4S pathway has been found to be quite slow, preventing this method of biodesulfurization from becoming commercially feasable. This is due to a number of limitations found in the native setup of the system. With synthetic biology approaches however, it is possible to optimize this pathway by making some alterations.
Our Approach
1) Find an organism
Plasmid was isolated(miniprepped) from and Rhodococcus. (I am not sure what is the name of the strain we got from Dr. Gieg). Using primers designed for amplification of each of dszA, dszB, dszC and hpaC these genes were isolated. The primers were designed in a way to add biobrick cut sites to genes. Afterwards, each gene was biobricked into a psb1c3 backbone. The next step was to construct the genes into an operon. However, before construction we had to mutate the enzymes. The promoter controlling the transcription of this pathway is repressed by sulphur containing compounds such as cysteine, methionine, and sulphate (Li et al. 1996). This problem is easily solved since our final construct will have a different promoter from the registry.
Replacing DszD with Hpac & Introducing Catalase
Based on the 4S pathway, FMNH2 is necessary for the functioning of dszC and dszA. A lack of FMNH2 slows down the DBT oxidation. A high concentration of FMNH2 on the other hand produces hydrogen peroxide (H2O2) that is lethal to cell in high concentrations (Gala´n et al. 2000). To overcome these problems two plans are implemented. First, hpaC gene is used instead of dszD which is simply another flavin reductase from Escheria coli W. It is shown that hpaC improves desulfurization by 7-10 times (Gala´n et al. 2000). Second, a catalase gene is used in the final construct to degrade any H2O2 species formed during FMNH2 recycling. hpaC gene was received in pUC18 plasmid from another university. (I am not exactly sure how David got this plasmid).
Mutagenesis: Biobrick Compatability and Increasing DszB Activity
dszA has four PstI cut sites. dszB has a PstI and a NotI cut site. dszC has two PstI cut sites. In order to be able to use these parts in construction, we had to eliminate these cut sites. Stratagene site directed mutagenesis was used for this purpose. During this procedure, two primers are designed for each mutation. Primers are complementary to the DNA strands except for one base pair. Therefore after the PCR, the whole plasmid is amplified with a point mutation. The base pair was chosen in a way so that it eliminates the cut site but the codon it belongs to still codes for the same amino acid (a silent mutation). We also used the same method to perform the Y63F mutagenesis in dszB (rationale will be explained). In this case the primers also introduce a point mutation. The point mutation causes the codon to code for Phe instead of Tyr. We also did this in a way so that the site of the mutation turns into a hpyAV cut site so we can confirm the mutagenesis by cutting the plamid with hpyAV.
Optimizing Gene Order
Rearranging the order of the genes in the operon is also shown to be beneficial in the efficiency of the 4S pathway. The natural order of genes in the operon is dszABC???. Our final construct, however, is dszBCA. This has multiple advantages over the natural operon. First of all the catalytic activity of DszA:DszB:DszC is 25:1:5 respectively. Therefore, the new arrangement produces more of the less active enzyme and less of the more active one by transcribing more of the less active enzyme which translates into more enzyme (Li et al. 2008). Second, the initiation codon of dszB overlaps with the termination codon of dszA in the natural operon which is also solved in the new construct. (we should explain why this is a problem i am not very clear on it apparently it causes translational coupling)
Final Constructs
 "
"