Team:Calgary/Project/OSCAR/CatecholDegradation
From 2012.igem.org


Hello! iGEM Calgary's wiki functions best with Javascript enabled, especially for mobile devices. We recommend that you enable Javascript on your device for the best wiki-viewing experience. Thanks!
Catechol Degradation

Catechol is a toxic compound found in tailings ponds that is a by-product of polyaromatic hydrocarbon metabolism (Vaillancourt et al., 2006, Schweigert et al., 2001)). The chemical properties of catechol allow it to react with biomolecules, causing cellular damage including DNA damage, enzyme inactivation and membrane uncoupling (Schweigert et al., 2001).
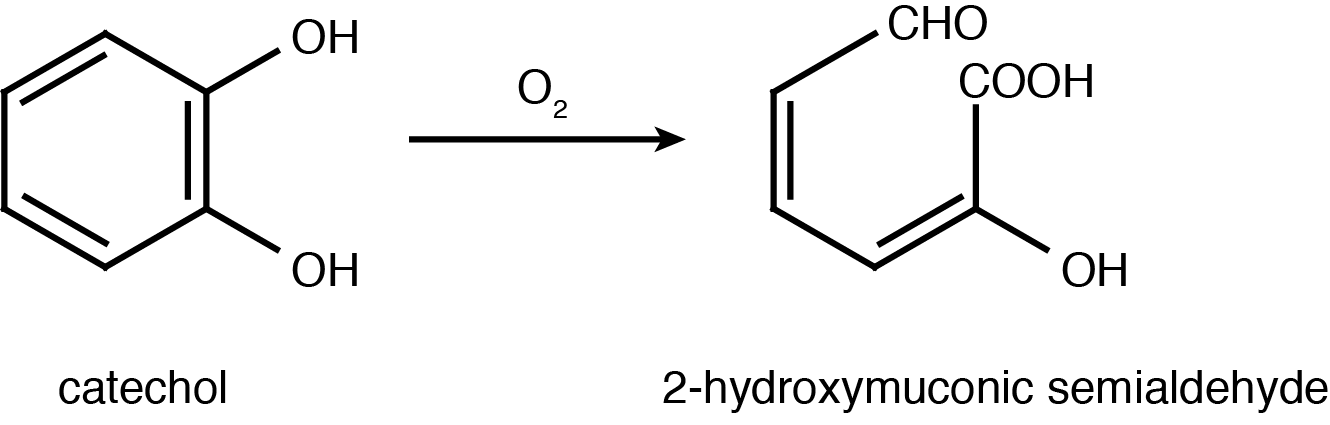
Catechol is characterized as having a benzene ring with two hydroxyl groups at the 2,3 position. It can be converted to 2-hydroxymuconic acid by the enzyme catechol 2,3-dioxygenase, encoded by the xylE gene on the Tol plasmid of Pseudomonas putida (Nakai et al., 1983).
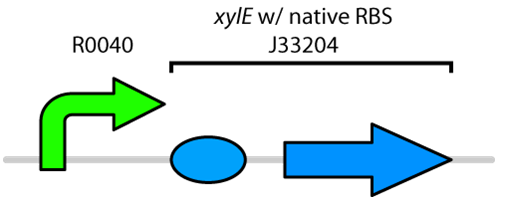
Currently the registry has two BioBricks available of xylE. One contained xylE with its native ribosome-binding site (BBa_J33204), while the other part contained xylE under the glucose-repressible promoter cstA (BBa_K118021). Given that E. coli is grown in the presence of glucose, we designed a new construct to keep xylE repressed by using the tetR promoter (BBa_R0040).
Catechol 2,3-dioxygenase is an extradiol dioxygenase which cleaves catechol adjacent to the two hydroxyl groups. When this occurs 2-hydroxymuconate semialdehyde is produced, which is yellow in colour. This change in colour allows for visual assay to assess the activity of XylE.
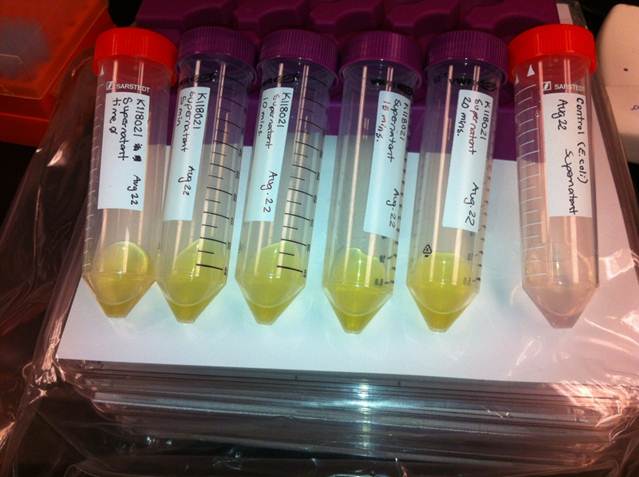
The visual assays were performed with E. coli cells transformed with (BBa_K118021) as well as with E. coli cells transformed with the newly constructed part (BBa_K902048) by bringing the supernatant of an overnight culture to a concentration of 0.1 M of catechol. When the part (BBa_K118021) was used, the pellet was first washed in M9-MM and centrifuged before catechol was added to the supernatant. This was necessary to avoid the glucose in the LB from repressing the cstA promoter (BBa_K118011). Catechol was added to the supernatant because the reaction takes place outside of the cell. Within minutes of the addition of catechol to the supernatant, the solution turned from the pale yellow of LB to a bright yellow. This was indicative that catechol was breaking down into 2-Hydroxymuconate semialdehyde, which was exactly what we expected! This assay was completed by following the protocol written by the 2008 Edinburgh iGEM team.
Converting Catechol into hydrocarbons?
After verifying that we could in fact degrade catechol into 2-hydroxymuconate semialdehyde using our xylE construct (BBa_J33204), we wondered if we could take this any further. What if we could convert this by-product page into hydrocarbons too? As catechol is the breakdown product of a number of different degradation pathways, this could be particularly useful.
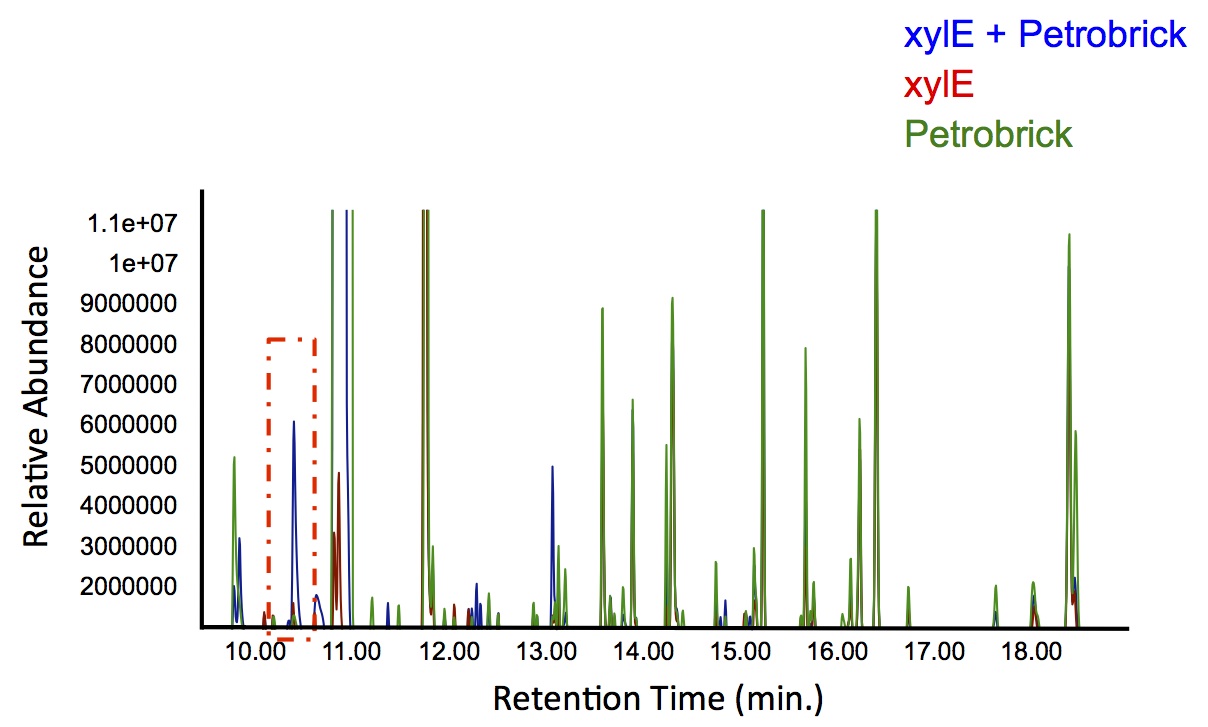
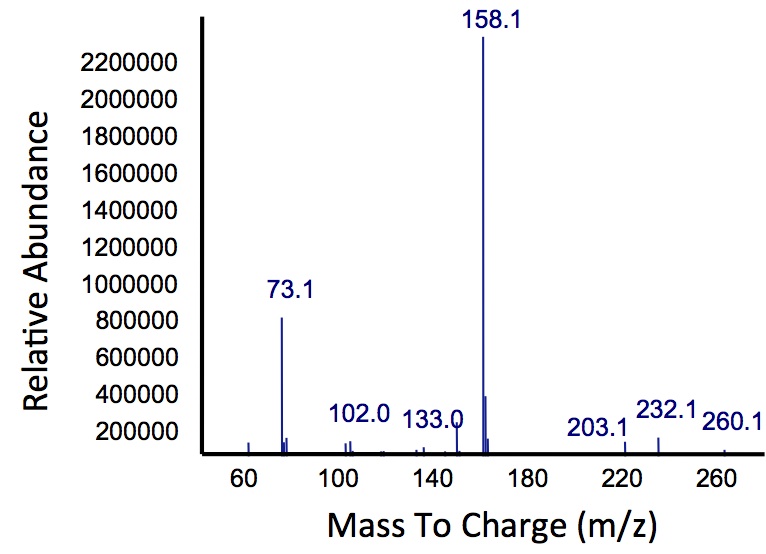
As 2-hydroxymuconate semialdehyde can be further metabolized to pyruvate and acetaldehyde (Harayama S et al., 1987), it seemed possible that these products could be routed into the fatty acid biosynthesis pathway and converted to alkanes using the PetroBrick or the OleT enzyme. Given that the Catechol 2,3-dioxygenase reaction is extracellular, it creates a possible scenario in which cells with the xylE construct could be co-cultured with Petrobrick-containing cells to cooperatively metabolise catechol into hydrocarbons.
In order to test this, we followed this protocol, where we co-cultured cells expressing our xylE construct with either E. coli cells expressing the PetroBrick, or Jeotgalicoccus sp. ATCC 8456 cells expressing OleT. in the presence of catechol.
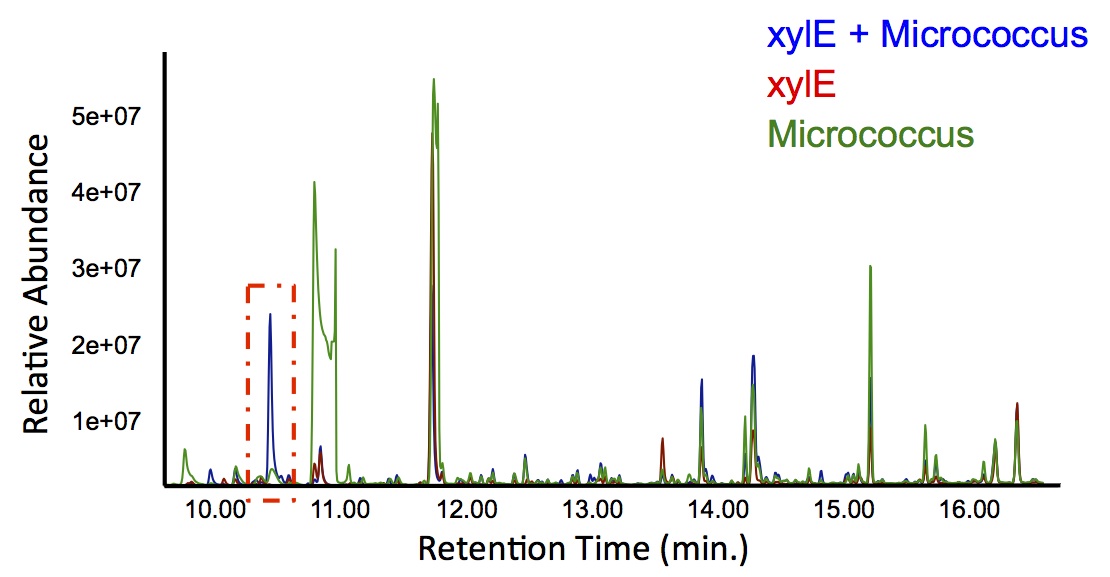
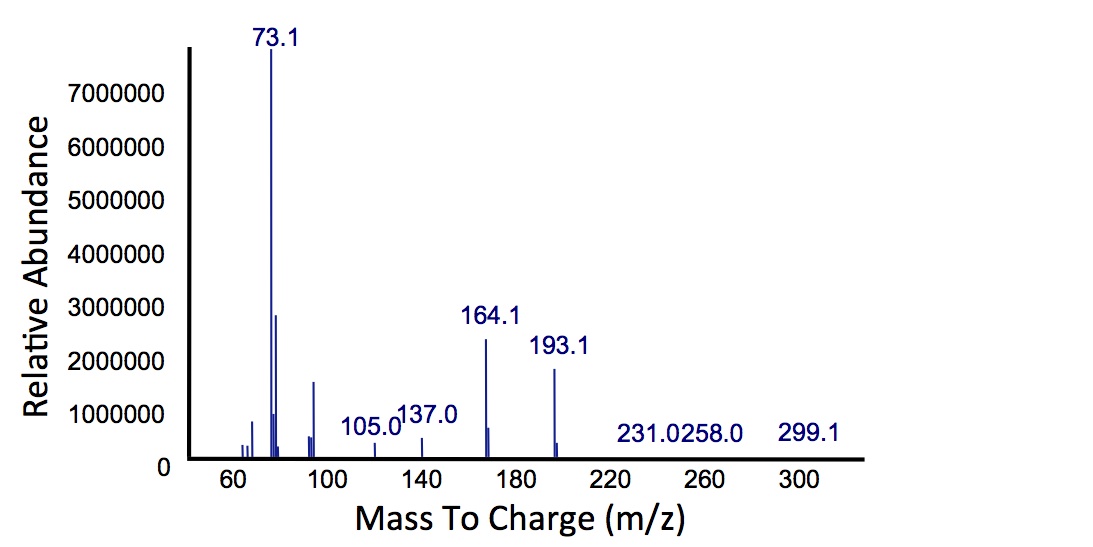
 "
"