Team:Calgary/Project/HumanPractices/Killswitch
From 2012.igem.org


Hello! iGEM Calgary's wiki functions best with Javascript enabled, especially for mobile devices. We recommend that you enable Javascript on your device for the best wiki-viewing experience. Thanks!
A Killswitch for Increased Security

Purpose:
The OSCAR component of our project aims to remediate toxins in the oil sands tailings ponds using synthetic bacteria. As we recognize in our project's safety practices(LINK), remediating tailings ponds creates the risk for our bacteria to escape into the environment, given the high volume of tailings water being circulated through our system. Although no evidence suggests that the OSCAR bacteria would proliferate or cause harm outside the bioreactor, industry leaders have voiced concerns to our team about synthetic biology's potential for environmental issues—safety is different in synthetic biology from more traditional engineering disciplines because bacteria are self-replicating entities. Thus, we cannot predict all possible consequences of OSCAR escaping the bioreactor.
To counter potential safety issues, we engineered a genetic-based containment mechanism into our bacteria. Our kill switch system is designed to destruct the organism's synthetic genes upon escape from the bioreactor, thereby lessening the possibility of OSCAR spreading beyond the bioreactor or horizontally transferring genes to other organisms. We hope that our proactive approach to safety in OSCAR will promote adoption of our synthetic biology system for remediation of tailings ponds in the conventional oil sands industry.
Our System:
Two basic elements comprise our active kill system: firstly, two nucleases are used to degrade the genome; and secondly, a regulatory platform is used to control these two kill genes. The nuclease enzymes ensure degradation of synthetic genes upon kill switch activation. Such a nuclease mechanism is superior to widely used lysis-based techniques which leave genetic material intact, allowing its release into the environment and potential horizontal gene transfer into other organisms. Our team submitted into the registry two novel kill enzymes (LINK). These are noteworthy because they are optimized for temperature conditions typical of tailing ponds. Furthermore, they cause finer degradation of the bacterial genome than existing nuclease mechanisms such as (SHOW ME ONE).
Of course, introducing nucleases is insufficient to control the spread of our organism and its genes. Our system also required a means of regulating the expression of these nucleases, thereby allowing the organism to function as intended while within the bioreactor environment. To fulfill this need, we developed four novel regulatory elements for registry submission, significant because they are tightly regulated—these elements reduce inadvertent expression of the kill genes compared to existing regulatory mechanisms. Three are of particular interest because they are riboswitches that regulate translation at the post-transcriptional stage of gene expression. Of the four we developed, two proved suitable for tailings pond use.
Justification of our approach
An apparent weakness of our system lies in the potential for the kill switch mechanism to mutate, rendering it ineffective and allowing the synthetic organism or its genetic material to escape into the environment. We have, however, taken several approaches to mitigate this risk. Firstly, we engineered redundancy into our system—with two kill genes, both would have to be rendered inoperable for the kill switch mechanism to malfunction. Knudsen et al. (1995) proposed that active kill switches containing a single kill element were subject to a mutation rate of 10^-6 per cell per generation, but that a second redundant kill gene reduces this value by two orders of magnitude. Secondly, the kill switch mechanism is, of course, only a failsafe measure for controlling our organism's spread. The primary means of preventing its escape is through the multiple layers of mechanical security provided by our bioreactor (LINK TO SAFETY). Only when these measures fail will the kill switch be required to function.
At first blush, an auxotrophic bacterial strain would seem a superior choice for our application. Such a strain would have a critical metabolic gene knocked out, requiring the organism be supplemented externally with an intermediate metabolite. As a mutation restoring the metabolite would be sufficiently complex as to be rendered improbable, an auxotrophic mechanism seems ideal. This system, however, is impractical due to its cost. Metabolic supplements are considerably more expensive than the glucose and metal ions that our system requires. As we learned in our discussions with industry leaders, cost is a paramount factor in permitting adoption at industrial scales, and so our system has a greater likelihood of being implemented outside the laboratory. More worrisome is that an auxotrophic control mechanism would kill the organism without degrading its genetic material, thereby making possible horizontal gene transfer to other organisms. As the overarching goal of our kill switch is not to kill the organism but to prevent the escape of its engineered genetic elements, our kill switch design is superior to one relying on auxotrophy.
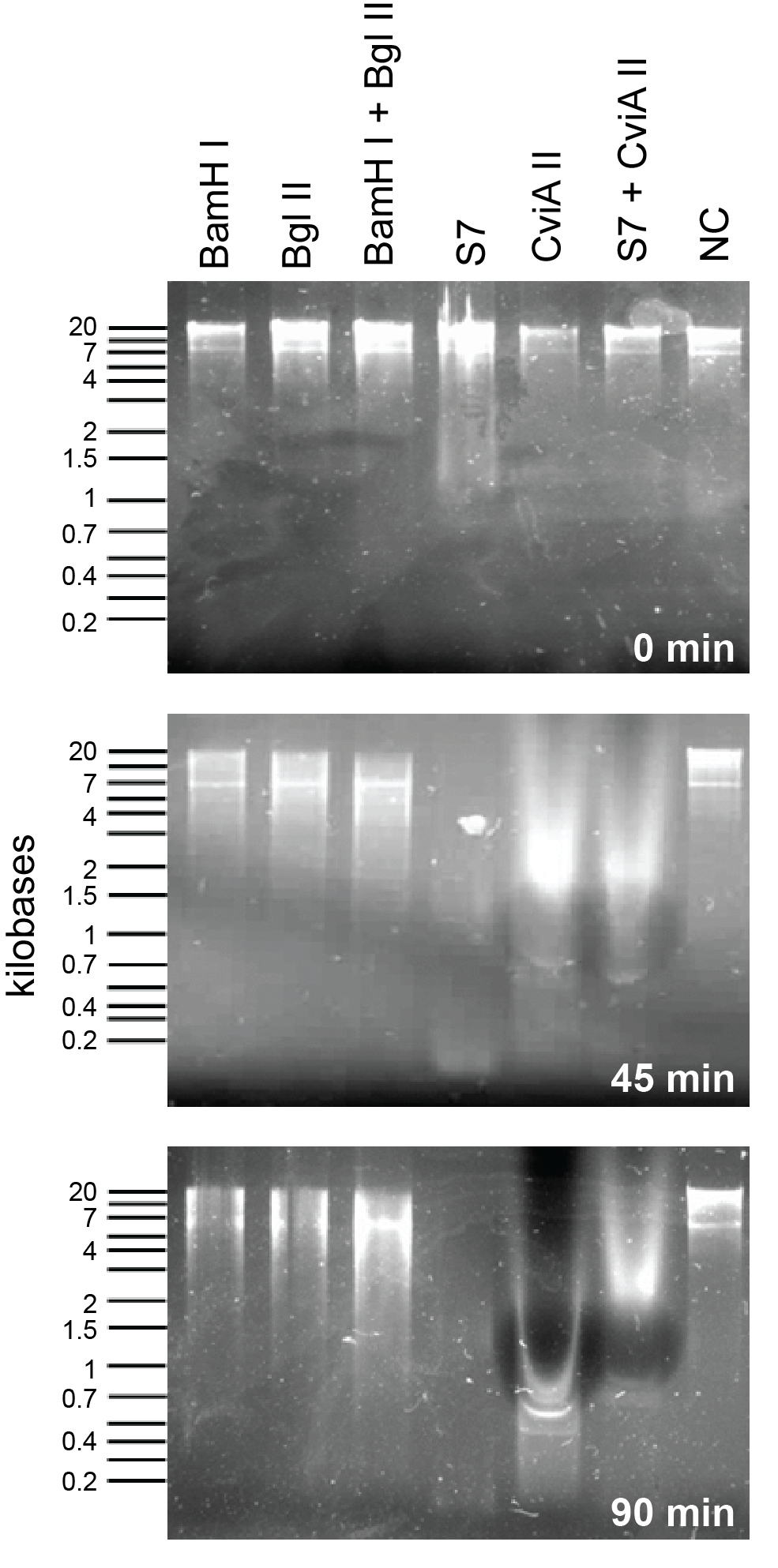
Nuclease assay to evaluate the nucleases present in the registry (BglII and BamHI):
To compare S7 and CviAII to the nucleases already present in the registry we did a nuclease assay with commercially available enzymes from New England Biolabs and an E. coli genomic prep. To see detailed protocol please link see here/link. As can be seen in Figure X, S7 starts acting almost immediately. Within 45 minutes both S7 and CviAII have chewed up the E. coli genome into small fragments whereas BamHI and BglII have sheared the genome into large fragments. Additionally, in 90 minutes, S7 and CviAII have sheared the genome into pieces <200 bp in size whereas there is no difference in the lanes with BglII and BamHI at 90 minutes compared to 45 minutes.
S7 micrococcal nuclease
S7 nuclease is native to Staphylococcus aureus. S. aureus uses this enzyme to destroy extracellular DNA when it infects humans. S7 has both endo and exonuclease activity. This enzyme has a preference for -AT rich regions as opposed to -GC rich regions. However, this enzyme digests the DNA into <200 bp fragments. Ideally this enzyme will be present both intracellularly and extracellularly. We synthesized this enzyme from IDT. However this came with a mutation which altered a lysine residue to an isoleucine thereby making the enzyme dysfunctional.
CviAII restriction enzyme
CviAII is a restriction endonuclease that was sourced from the Chlorella virus PBCV-1 (REF PAPER). Our team selected this enzyme for three reasons.
Firstly, this enzyme recognizes the small four-base restriction site CATG wherein it cuts a staggered end between the A and C on the forward and reverse strands. This is advantageous for the design of our system because of the frequency of this short cut site in the E. coli genome. As opposed to the six base cutter BamHI system submitted by the 2007 Berkely team (BBa_I716462), the CviAII restriction site is to be 16 times more prevalent in the E. coli genome, which translates in finder degradation of the genetic material.
Secondly, the CATG cut site has the probability of being present in start codons of one quarter of genes in the E. coli genome. As such, coding genes will preferentially be selected with activation of CviAII; at this point, the exonuclease activity of S7 micrococcal nuclease can complete degradation of the gene element. Additionally, CviAII is able to cut Dam and Dcm methylated sites in the E. coli genome, and this translates into decreased selectivity of the enzyme.
Finally, the optimum temperature for CviAII activity is 23 degrees Celsius (REF PAPER). This value is relatively low and better suited to operation the cooler tailings water compared to other systems in the registry. For example, the 2007 Berkely BamHI system is optimized for 37 degrees Celsius and thus would be non-functional the tailings ponds. FIND SOME DATA OF TAILING WATER TEMP.
Regulation of our kill genes:
We have explored four different systems in our project. All of these systems fall under the umbrella of inducible kill gene systems. They are: Glucose repressible system, magnesium repressible system , manganese inducible system and the molybdate repressible system.
Molybdate co-factor protein regulation
 "
"