Team:University College London/LabBook/Week11
From 2012.igem.org



Contents |
(11.1) Monday 20.08
Aim: To test how the amount of nuclease produced by W3110 E. coli cells changes over time
Methods: (to make in a protocol format please
For Wnu cell line which has native secreted nuclease activity
1. Prepare 11-16 plates (10ml LBAgar+10ul AMP +10 ul 1M IPTG). IPTGinduces the lac promoter which in turnactivates the transcription of nuclease.
2. Streak cells onto all plates at the same time
3. Incubate at 37°C
4. Applyhydrchloric acid (HCL) to the first plate before putting in the incubator (set time as zero)
5. Take a second reading after four hours, followed by six readingsevery 3 hours, and a final three readings every two hours.
6. When the reading is taken, observe the following:
a) Diameter of the colony (once the diameter of the colony is measured, pick the colony and put it to grow in LB for nine hours)
b) Diameter of the halo that is achieved once HCL is applied
c) OD from a)
d) Estimate of the depth of the colony on the agar plate
For BL21 cell line that has been modified to contain nuclease
7. Prepare 11-16 plates (LB Agar + CMP)
8. Streak cells onto all plates at the same time
9. Incubate at 37°C
10. Apply HCL to the first plate before putting in the incubator (set time as zero)
11. Take a second reading after four hours, followed by six readings every 3 hours, and a final three readings every two hours.
12. When the reading is taken, observe the following:
a. Diameter of the colony (once the diameter of the colony is measured, pick the colony and put it to grow in LB for nine hours)
b. Diameter of the halo that is achieved once HCL is applied
c. OD from a)
d. Estimate of the depth of the colony on the agar plate
Results: The following table shows readings of the colony and halo diameters, plus the OD, taken at different times over 28 hours.
| Date | Time | Colony diameter | Halo diameter | Absorbance at 600 OD |
|---|---|---|---|---|
| 20.08.2012 | 12.30pm | 0 | 0 | 0 |
| 20.08.2012 | 16.30pm | 0 | 0 | 0 |
| 20.08.2012 | 19.30 pm | 0 | 0 | 0 |
| 20.08.2012 | 22.30 pm | 0.5mm | 1mm | 0.001 |
| 21.08.2012 | 01.30 am | 1mm | 3mm | 0.142 |
| 21.08.2012 | 04.30 am | 1.5mm | 7mm | 0.228 |
| 21.08.2012 | 07.30 am | 1.5mm | 7.5mm | 0.252 |
| 21.08.2012 | 10.30am | 2.5mm | 11mm | 0.522 |
| 21.08.2012 | 12.30pm | 3mm | 13mm | 0.749 |
| 21.08.2012 | 14.30pm | 3.5mm | 14.5mm | 0.844 |
| 21.08.2012 | 16.30pm | 4mm | 16mm | 0.937 |
Conclusion: From the data collected, it is clear that colony size correlates with the nuclease produced, which is represented by the diameter of the halo. Readings indicated that both are proportionally related to time. The second run of the experiment will be done in the coming weeks so as to obtain data replicates in order to validate data (See Week 13).

(11.2) Tuesday 21.08
Aim: To make chemically competent W3110 E. coli Cells – Day 1
Methods: Day 1 protocol. See previous experiments.
Wednesday 22.08
Aim: To make chemically competent W3110 E. coli Cells – Day 2
Methods: Day 2 protocol. See previous experiments.
Thursday 23.08
Aim: To make chemically competent W3110 E. coli Cells – Day 3
Methods: Day 2 protocol. See previous experiments.

(11.3) Tuesday 21.08
Aim: To carry out PCR clean-up to purify the Laccase gene and Curli cluster of genes achieved from PCR carried out
Method:
Please refer to previous protocols
The following table shows the gene which was extracted from PCR and the primers which were used for the PCR.
| Gene | Primers used |
|---|---|
| Curli | CF1 - C2R |
| Curli | CF3 – C4R |
| Curli | CF1 – C4R |
| Curli | CF3 – C2R |
| Laccase | LFONE - LR1 |
| Laccase | LFTWO - REVLF2 |
| Laccase | LR1 – LFTWO |
| IrrE | STF1 – ST2R |
| IrrE | STF3 – ST4R |
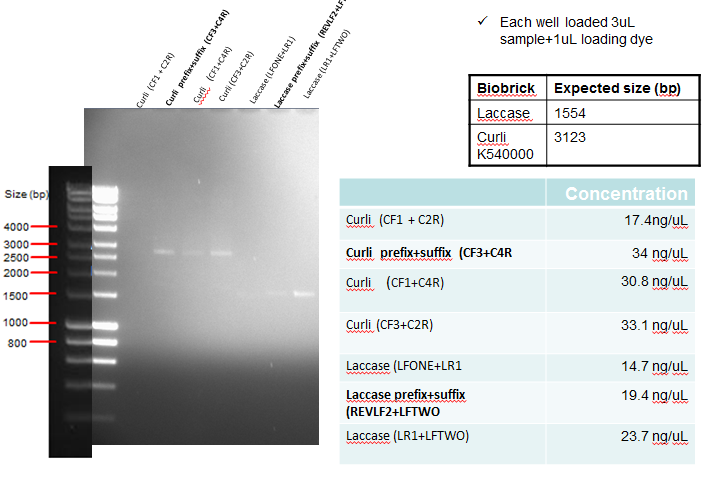
Results: The following diagram shows the results of a gel run to confirm plasmid identity of Curli and Laccase after PCR clean-up, with each well having 3ul of PCR purified product. In addition the table on the bottom right shows the concentration of the purified products. The samples in bold below represent the PCR reactions which where considered successful, and hence taken forward for ligation.
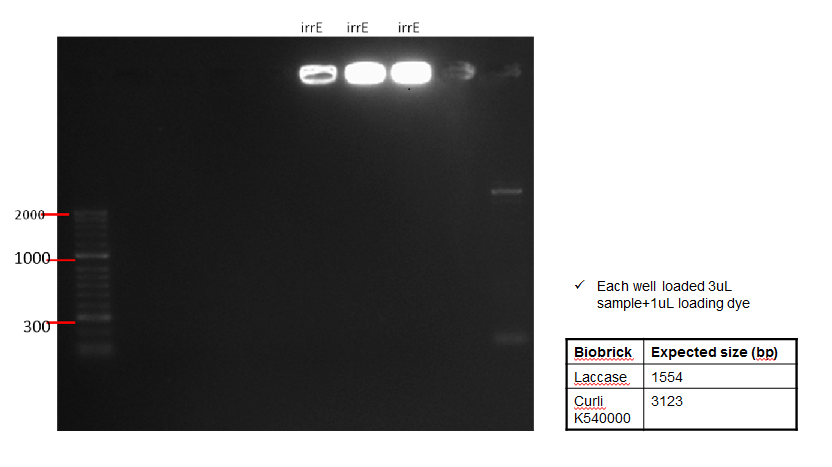
The following table shows a gel run with sampled of the purified IrrE gene. As can be seen from the gel, the purified material remained in the well, indicating that PCR clean-up was not successful.
Conclusion: Since PCR was unsuccessful, this was repetead (See Week 12)

(11.4) Wednesday 22.08
Aim: To carry out a preparative digest on the tetracycline plasmid backbone, cobalt curli, and terminator and nuclease in preparation of ligation.
Methods:
Please refer to previous runs
Aim: To carry out a ligation of cobalt curli and the terminator sequence and another ligation of the nuclease and terminator sequence, in preparation of further. This was done by ligating the terminator sequence to the curli cluster of gene through a EcoRI-PstI ligation, and another ligation of the terminator to nuclease through a EcoRI-PstI ligation.
Once success of ligation is confirmed, two subsequent ligations will be carried out to attach the ligated sequences to the promoter and rbs, and eventually the backbone.
Methods:
Please refer to previous runs
Aim: To transform W3110 cells with the ligated sequences from 3.
Methods:
Please refer to previous runs
The following table shows the number of plates with 10ul and 90ul of transformation product:
Ligation Number of plates with 10ul volume of transformation product Number of plates with 90ul volume of transformation product
| Curli + terminator | 3 | 3 |
|---|---|---|
| Nuclease + terminator | 3 | 3 |
In addition, three controls were used; one agar plate with no antibiotic to act as a positive control, one agar plate with ampicillin instead of nuclease to act as a negative control and a final agar plate with chloramphenicol instead of curli to act as a negative control.
Thursday 23.08
Aim: To check whether transformation carried out using ligated sequences was successful.
Results:
A large number of clear, isolated colonies were observed on all transformation plates. The positive control showed extensive growth, while the negative controls remained clear, indicating a successful transformation..
Conclusion: Since growth was as expected, colonies were picked for inoculation, as will be explained in the next step
Aim: To inoculate colonies from the transformation plates in preparation for purification and use in further ligations.
Methods:
The following table shows the number of colonies picked:
| Transformed ligation products | Number of colonies |
|---|---|
| Curli + terminator | 4 |
| Nuclease + terminator | 3 |
In addition, three controls were used. One consisted of LB, another of LB + Amp and a third of LB + CMP.
Friday 24.08
Aim: To purify ligated gene+terminator DNA using inoculated colonies from the ligation transformation plates
Results: No growth was noted in the falcon tubes inoculated with colonies.
Conclusion:
The purification protocol was not followed, and instead, a repeat of the transformation was carried out as will be explained next.
Aim: To repeat the transformation using the ligation samples.
Methods:
Please refer to previous runs
| Ligation | Number of plates with 10ul volume of transformation product | Number of plates with 90ul volume of transformation product |
|---|---|---|
| Curli + terminator | 3 | 3 |
| Nuclease + terminator | 3 | 3 |
 "
"