Team:Calgary/Notebook/Protocols/mutagenesis
From 2012.igem.org


Hello! iGEM Calgary's wiki functions best with Javascript enabled, especially for mobile devices. We recommend that you enable Javascript on your device for the best wiki-viewing experience. Thanks!
Site-Directed Mutagenesis
We used two types of mutagenesis protocols. The first method introduces a silent mutation to eliminate a biobrick cut site (EcoRI, NotI, XbaI,SpeI, or PstI) in the gene of interest; the second method introduces a mutation in the gene to change an amino acid in the final protein product.
To introduce a silent mutation (first method), primers were designed in order to change a base pair in a codon without affecting the amino acid sequence (a silent mutation).
In the second method, one base pair in a codon so the codon now codes for a (new) desired amino acid while also introducing a non-biobrick restriction site in the gene. Successfully mutated genes can be screened for by cutting with the particular restriction enzyme whose site has been created. In some cases, it mmay be necessary to mutate more than one base pair to create the restriction site.
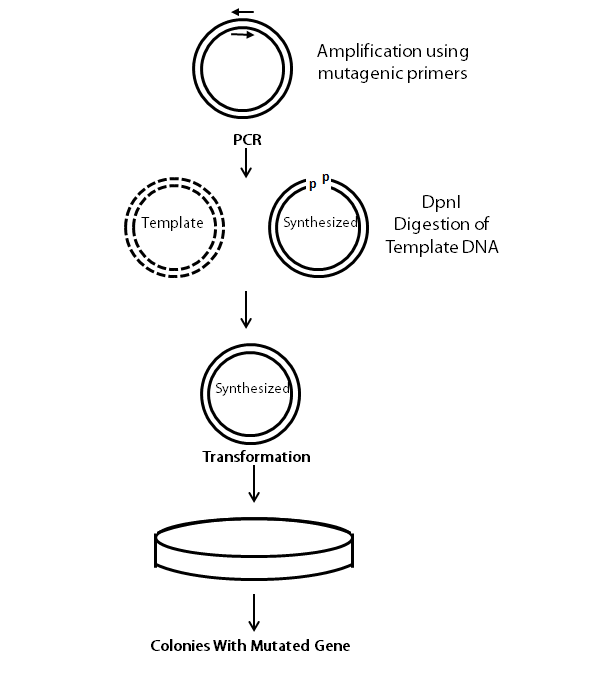
The following flow chart provides a visual guideline as to how the entire is conducted.
Primer Design
(Modified from Stratagene's QuikChange Site-Directed Mutagenesis Kit)
Two primers (forward and reverse) are designed to be complementary to the target gene sequences except for the base pair to be mutated. The primers must adhere to the following criteria:
- The mutation must be in the middle of the primer with 10-15bp on each side.
- The primers must be between 25-45bp.
- The primers must introduce the same mutation.
- The primers must have a GC content of more than 40% and they must end on each side with at least a G or C.
- Primers must have a Tm of at least 78°C based on the following formula:
Tm = 81.5 + 0.41(%GC) - 675/N - %mismatch
PCR reaction
During the PCR reaction, the mutagenesis primers bind to the dissociated DNA strands of the plasmid, and the DNA polymerase synthesizes the whole plasmid. To find out the right proportions of primers to plasmid DNA, primer concentrations is kept in excess and different concentrations of the DNA template are used to find the optimum concentration. We used the KAPA HiFi PCR Kit. The PCR reaction is set up as follows:
| KAPA 5x Fidelity buffer: | 5 µL |
| KAPA DNTP mix: | 0.75 µL |
| Forward primer: | 0.75 µL |
| Reverse primer: | 0.75 µL |
| Plasmid: | Try different concentrations (5ng, 20ng, 50ng) |
| KAPA HiFi DNA Polymerase: | 0.5 µL |
| MilliQ water: | up to 25 µL |
Transformation
For confirmation, 10 µL of the PCR reaction is run on a gel. If amplification is observed, 1 µL of DpnI enzyme is added to the remaining PCR reaction (15µL) and incubated at 37°C for one hour. DpnI enzyme degrades the methylated parental DNA. Since, synthesized DNA from PCR is not methylated, DpnI cannot digest the PCR products.
Afterwards, the digested PCR prodcuts are transformed into E. coli. 1µL of the PCR product is added to 50 µL of chemically competent cells. Follow the transformation procedure provided.
Screening
Plasmids are purified from overnight cultures of transformed colonies, and digested with the appropriate enzyme (depending on site introduced initially) for verification. The plasmid without the desired mutation is digested as a negative control. Digested products are run on a gel in order to detect the presence of the desired mutation. If the sizes (in kb) of the bands are as expected, the mutagenesis should be successful. The plasmid is sent for sequencing for confirmation as in some instances random mutations or insertions happen (e.g. insertions near the primer binding sites may occur).
 "
"