Team:Calgary/Project/FRED/Modelling
From 2012.igem.org
| Line 35: | Line 35: | ||
</html> | </html> | ||
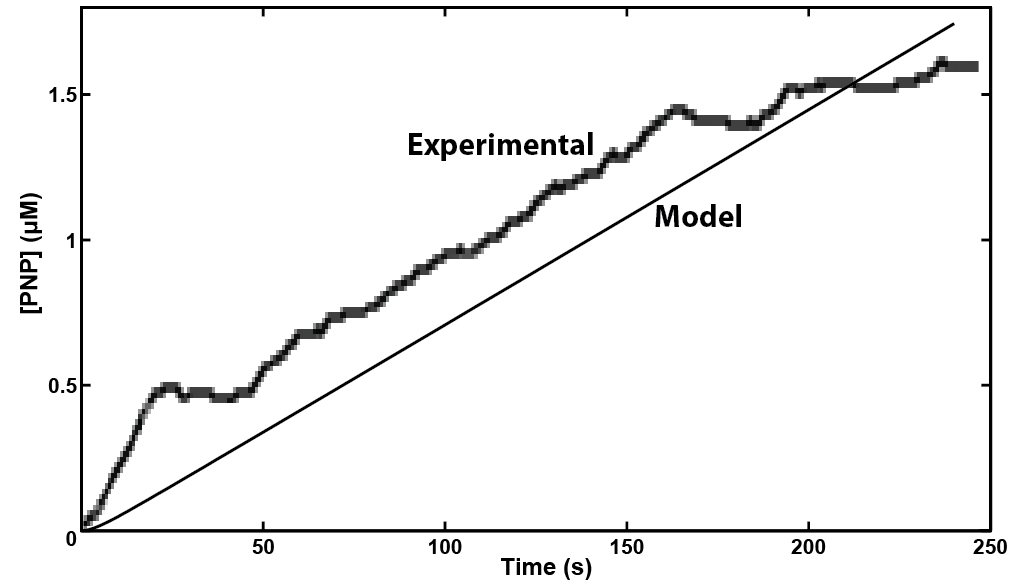
| - | [[File: | + | [[File:Calgary Screen Shot 2012-10-03 at 9.14.48 AM.png|thumb|600px|center|Figure 3: Experimental and modelling results for the <i>uidA</i> gene to cause cleavage of PNPG into PNP. Both results show that a detectable level of PNP is reached in under 4 minutes.]] |
<html> | <html> | ||
Revision as of 15:21, 3 October 2012


Hello! iGEM Calgary's wiki functions best with Javascript enabled, especially for mobile devices. We recommend that you enable Javascript on your device for the best wiki-viewing experience. Thanks!
A Kinetic Model in MATLAB

The creation of our electrochemical sensor presented us with an interesting question: How fast would the sensing response be? Instead of trying many timecourses in the lab, we turned to modelling to give us an answer. We have created a kinetic model for one of our systems, the uidA system, to give us an estimate of the time needed to observe a result.
How did we build it?
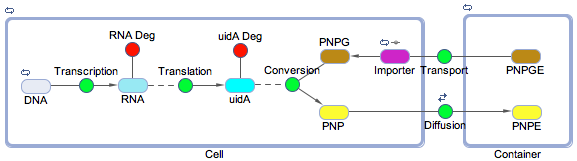
Using the SimBiology toolbox for MATLAB we created a framework of the electrochemical reporter system. We started at the genetic level, modelling transcription, translation, and the enzyme catalysis. We also included the rate of transport of the substrate into the cell and the diffusion of the product out of the cell. The degradation reactions for the RNA transcript and the enzyme were also included. This reaction scheme is shown below in Figure 1.
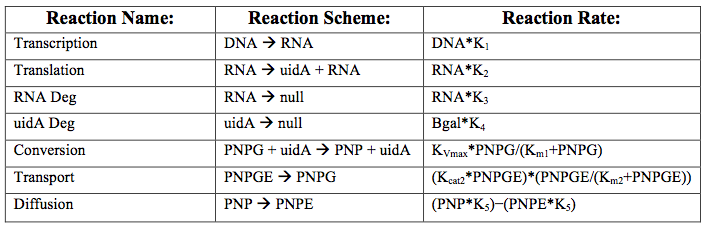
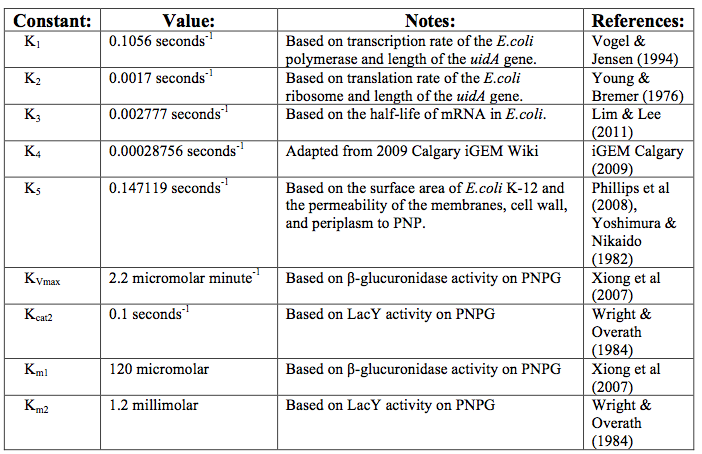
After creating the basic framework for the model we needed to create mathematical equations for each reaction with appropriate rate constants. These equations and the corresponding values are shown below in tables 1 and 2.
What did it show?
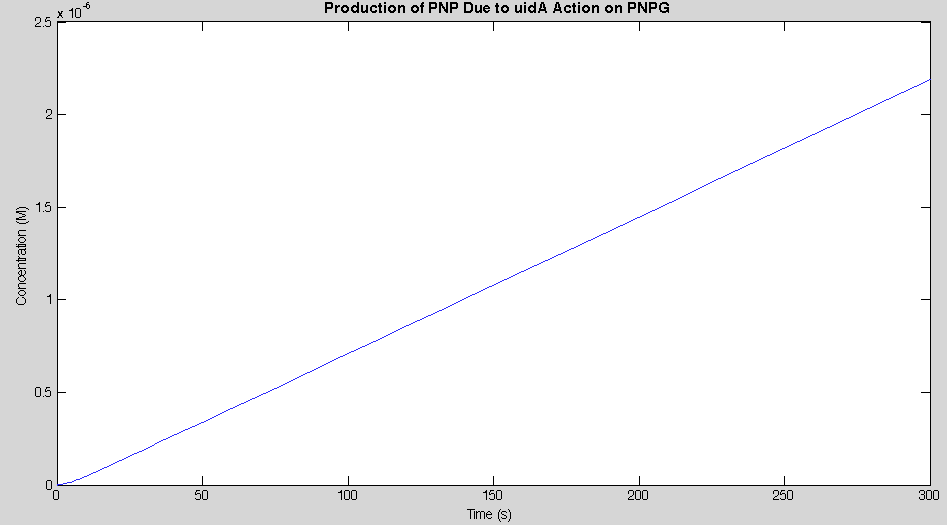
Before running the model we needed to decide what an appropriate endpoint would be. As we were trying to determine how quickly we could get a result we chose 2µM as our endpoint. This was chosen because initial electrochemical testing of PNP showed strong responses at concentrations as low as this. With our endpoint chosen the model was run and the data is shown below in figure 2.
Our model showed that under strong expression we would be able to see a response in under 4 minutes. This data was then used to design an experiment that could be performed in the lab to verify the model. A uidA circuit was induced in the lab and the subsequent cleavage of PNPG into PNP was detected electrochemically at 1.6V vs the Reduction of Hydrogen Electrode. The data from that test is shown below.
Future plans
There are ways that this model could be improved for future uses. One main avenue for further exploration is that of modelling the remaining two electrochemical systems (lacZ and bglX). The system modeled here was chosen because of the availability of data for the compound, both electrochemically and enzymatically. A second avenue for improvement would be to model varying promoter strengths to see how dramatically the timecourse would change over varying concentrations of an inducer. This could be expanded upon to predict appropriate promoters for a final system.
This model has shown us a timecourse and successfully informed the wetlab experiments. The iterative process of modelling and experimenting will give future versions of this model more data to use and increase its accuracy.
 "
"