Team:Calgary/Project/OSCAR/CatecholDegradation
From 2012.igem.org
| Line 6: | Line 6: | ||
<p><b>****This section needs work. Why are we degrading catechol? What part did we use? What is the number?</b></p> | <p><b>****This section needs work. Why are we degrading catechol? What part did we use? What is the number?</b></p> | ||
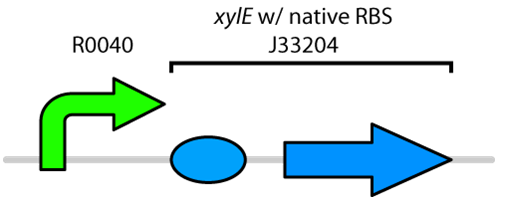
| - | <p>Catechol is a toxic compound found in tailings ponds that is a by-product of polyaromatic hydrocarbon metabolism. Catechol is toxic to a wide range of organisms from microorganisms to mammals (Schweigert <i>et al.</i>, 2001). The chemical properties of catechol allow it to react with biomolecules like DNA, proteins and membranes (Schweigert <i>et al.</i>, 2001). These interactions can cause serious damage including DNA breakage, enzyme inactivation and membrane uncoupling (Schweigert <i>et al.</i>, 2001). Catechol can be degraded by the enzyme catechol 2,3-dioxygenase encoded by the <i>xylE</i> gene on the Tol plasmid of <i>Pseudomonas putida</i> (Nakai <i>et al.</i>, 1983). The current iGEM Part repository has two BioBricks available of <i>xylE</i>. One contained <i>XylE</i> with its native ribosome-binding site (part: <a href=http://partsregistry.org/Part:BBa_J33204>J33204</a>), while the other part contained <i>XylE</i> under the glucose-repressible promoter cstA (Part: <a href=http://partsregistry.org/Part:BBa_K118021>K118021</a>). Given that <i>E. coli</i> is grown in the presence of glucose, we designed a new construct to keep <i>XylE</i> repressed by using the TetR promoter (Part:<a href= http://partsregistry.org/Part:BBa_R0040>R0040</a>).</p> | + | <p>Catechol is a toxic compound found in tailings ponds that is a by-product of polyaromatic hydrocarbon metabolism (Vaillancourt <i>et al.</i>, 2006). Catechol is toxic to a wide range of organisms from microorganisms to mammals (Schweigert <i>et al.</i>, 2001). The chemical properties of catechol allow it to react with biomolecules like DNA, proteins and membranes (Schweigert <i>et al.</i>, 2001). These interactions can cause serious damage including DNA breakage, enzyme inactivation and membrane uncoupling (Schweigert <i>et al.</i>, 2001). Catechol can be degraded by the enzyme catechol 2,3-dioxygenase encoded by the <i>xylE</i> gene on the Tol plasmid of <i>Pseudomonas putida</i> (Nakai <i>et al.</i>, 1983). The current iGEM Part repository has two BioBricks available of <i>xylE</i>. One contained <i>XylE</i> with its native ribosome-binding site (part: <a href=http://partsregistry.org/Part:BBa_J33204>J33204</a>), while the other part contained <i>XylE</i> under the glucose-repressible promoter cstA (Part: <a href=http://partsregistry.org/Part:BBa_K118021>K118021</a>). Given that <i>E. coli</i> is grown in the presence of glucose, we designed a new construct to keep <i>XylE</i> repressed by using the TetR promoter (Part:<a href= http://partsregistry.org/Part:BBa_R0040>R0040</a>).</p> |
<h3></h3> | <h3></h3> | ||
Revision as of 09:00, 3 October 2012


Hello! iGEM Calgary's wiki functions best with Javascript enabled, especially for mobile devices. We recommend that you enable Javascript on your device for the best wiki-viewing experience. Thanks!
Catechol Degradation

****This section needs work. Why are we degrading catechol? What part did we use? What is the number?
Catechol is a toxic compound found in tailings ponds that is a by-product of polyaromatic hydrocarbon metabolism (Vaillancourt et al., 2006). Catechol is toxic to a wide range of organisms from microorganisms to mammals (Schweigert et al., 2001). The chemical properties of catechol allow it to react with biomolecules like DNA, proteins and membranes (Schweigert et al., 2001). These interactions can cause serious damage including DNA breakage, enzyme inactivation and membrane uncoupling (Schweigert et al., 2001). Catechol can be degraded by the enzyme catechol 2,3-dioxygenase encoded by the xylE gene on the Tol plasmid of Pseudomonas putida (Nakai et al., 1983). The current iGEM Part repository has two BioBricks available of xylE. One contained XylE with its native ribosome-binding site (part: J33204), while the other part contained XylE under the glucose-repressible promoter cstA (Part: K118021). Given that E. coli is grown in the presence of glucose, we designed a new construct to keep XylE repressed by using the TetR promoter (Part:R0040).
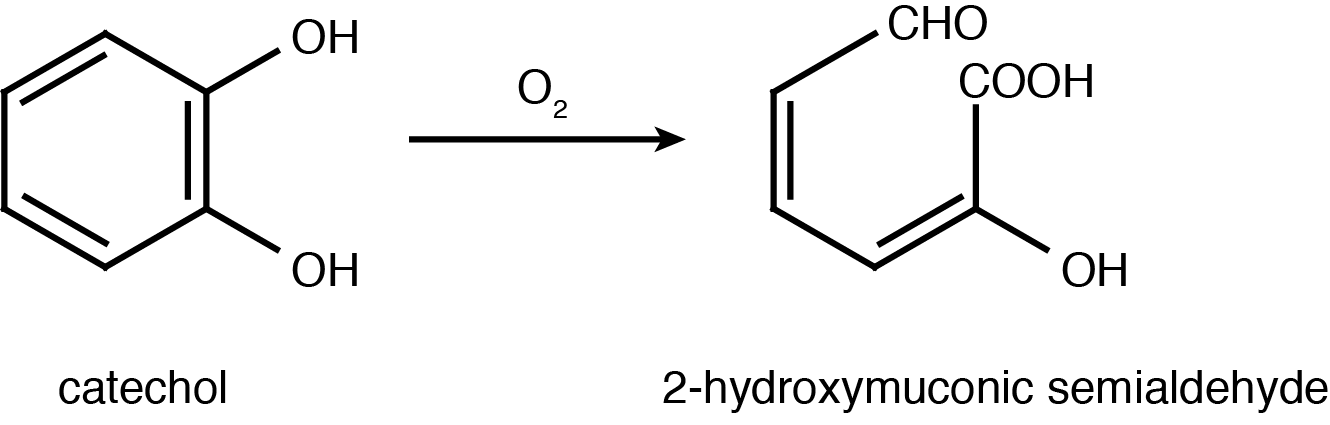
Catechol 2,3-dioxygenase is an extradiol dioxygenase which cleaves catechol adjacent to the two hydroxyl groups. When this occurs 2-hydroxymuconate semialdehyde is produced, which is yellow in colour. This change in colour allows for visual assay to assess the activity of XlyE.
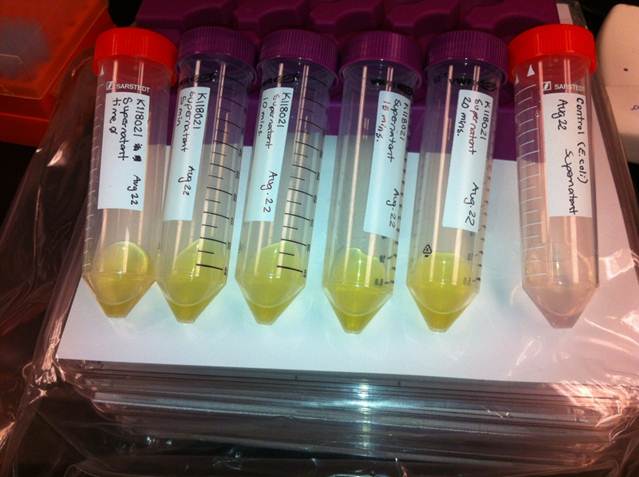
The visual assays were performed with E.coli cells transformed with K118021 as well as with E.coli cells transformed with the newly constructed part (K902048) by bringing the supernatant of an overnight culture to a concentration of 0.1 M of catechol. When the part K118021 was used the pellet was first washed in M9-MM and spun down before catechol was added to the supernatant. This was done to avoid the glucose in the LB from repressing the cstA promoter (K118011). The catechol was added to the supernatant because the reaction takes place outside of the cell. Within minutes of the addition of catechol to the supernatant, the solution turned from the pale yellow of LB to a bright yellow. This assay was completed by following the previous assay done by the 2008 Edinburgh iGEM team.
 "
"