Team:Calgary/Project/OSCAR/Denitrogenation
From 2012.igem.org
| Line 35: | Line 35: | ||
<h2>Our Constructs</h2> | <h2>Our Constructs</h2> | ||
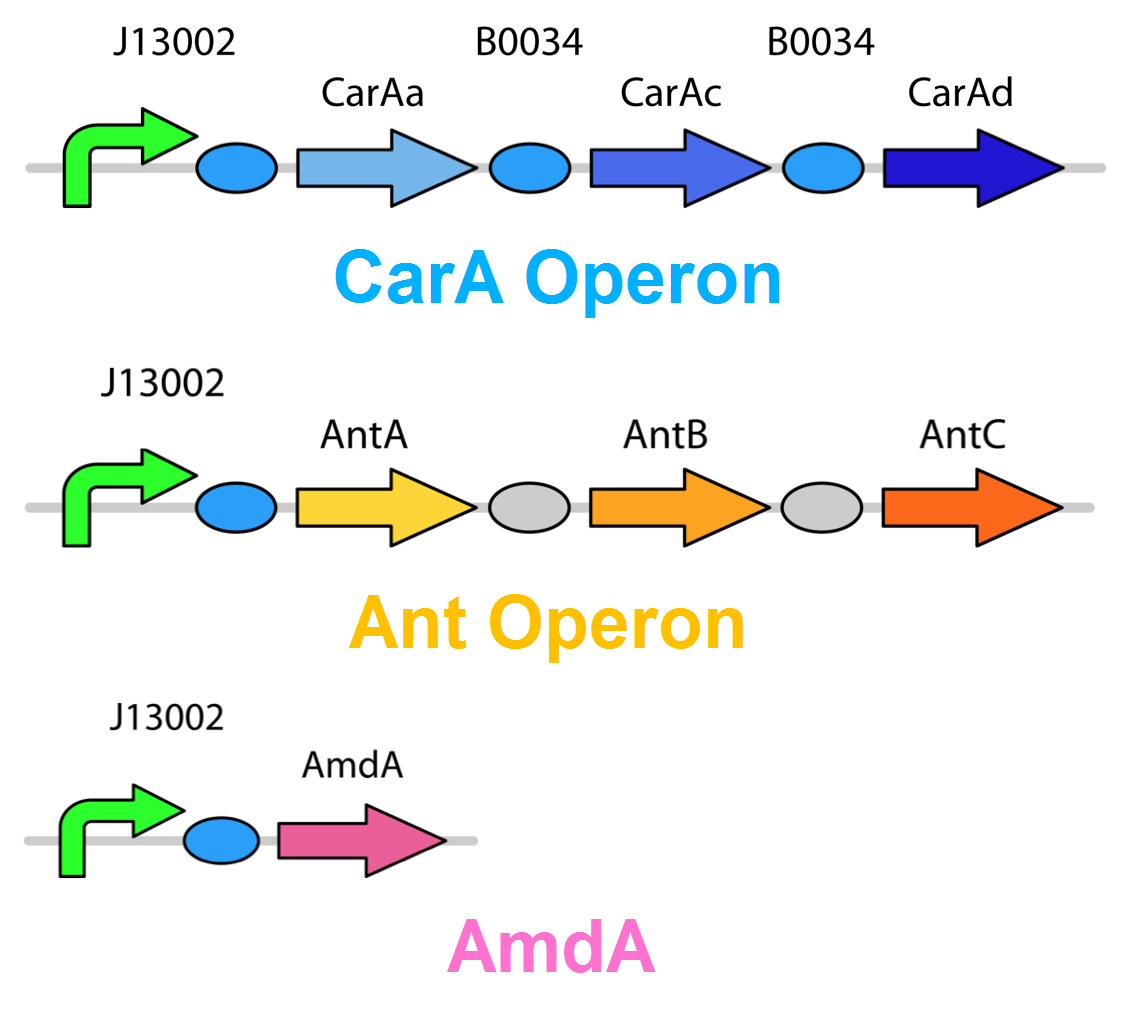
| - | <p>The <i>carA</i> genes have been biobricked individually into an operon under the control of a TetR promoter (BBa_J13002), with ribosome binding sites (BBa_B0034) inserted in front of each gene. The <i>carAd</i> gene was given a silent mutation to eliminate an illegal NotI site in the native sequence. The <i>ant</i> genes were biobricked as a whole operon with native ribosome binding sites in between each gene, also downstream of a TetR promoter. The <a href="http://partsregistry.org/Part:BBa_K902035"><i>AmdA</i></a> gene was also biobricked downstream of a TetR promoter and ribosome binding site. </p> | + | <p>The <i>carA</i> genes have been biobricked individually into an operon under the control of a TetR promoter (<a href="http://partsregistry.org/Part:BBa_J13002">BBa_J13002</a>), with ribosome binding sites (BBa_B0034) inserted in front of each gene. The <i>carAd</i> gene was given a silent mutation to eliminate an illegal NotI site in the native sequence. The <i>ant</i> genes were biobricked as a whole operon with native ribosome binding sites in between each gene, also downstream of a TetR promoter. The <a href="http://partsregistry.org/Part:BBa_K902035"><i>AmdA</i></a> gene was also biobricked downstream of a TetR promoter and ribosome binding site. </p> |
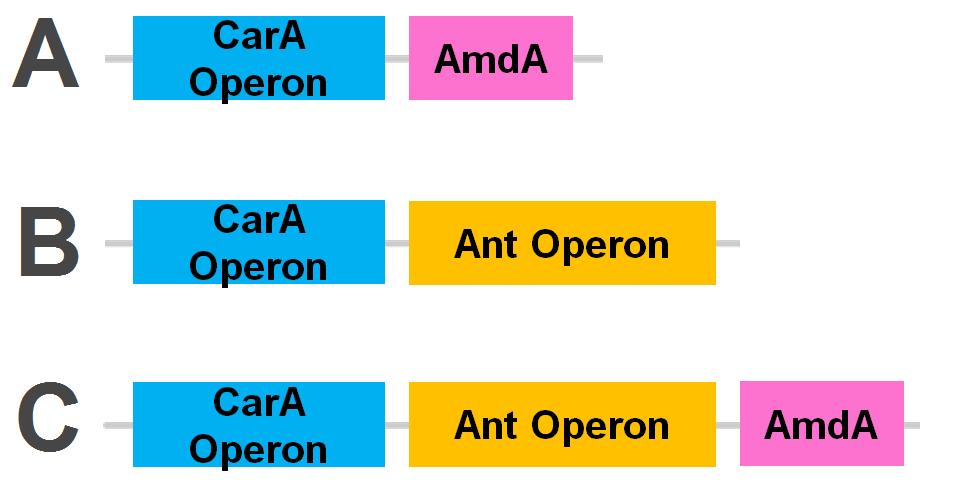
</html>[[File:Carbazole degradation constructs calgary12.png|thumb|420px|center|Genetic circuits for nitrogen removal. 2 operons and an additional gene have been biobricked together under 1 TetR promoter each.]]<html> | </html>[[File:Carbazole degradation constructs calgary12.png|thumb|420px|center|Genetic circuits for nitrogen removal. 2 operons and an additional gene have been biobricked together under 1 TetR promoter each.]]<html> | ||
Revision as of 06:18, 3 October 2012


Hello! iGEM Calgary's wiki functions best with Javascript enabled, especially for mobile devices. We recommend that you enable Javascript on your device for the best wiki-viewing experience. Thanks!
Denitrogenation

Why Focus On Nitrogen Bioremediation?
The removal of nitrogen heterocycles from the petroleum tailings ponds is important for both of the main goals of the OSCAR project. Since nitrogen containing compounds lower the quality and stability of fuel (Katzer & Sivasubramanian, 1979) , their presence would be detrimental to the usefulness of the hydrocarbons OSCAR could create. The presence of nitrogen compounds in our products also hinders our environmental goals as they have been shown to undergo radical changes, yielding highly genotoxic byproducts (Xu et al., 2006). The combination of these two factors makes the removal of nitrogen very important to the success of OSCAR.
Our Model Compounds
Carbazole was chosen as the model compound to study nitrogen containing heterocycles as it accounts for about 70% of nitrogen by mass in petroleum tailings ponds, as well as the fact that it is considered one of the more difficult to degrade (Morales, 2010). A pathway that is capable of degrading carbazole could also have the ability to degrade many other types of nitrogen containing compounds found in the tailings ponds. Although the genes in our pathway are mostly designed for carbazole degradation, 3 other compounds have also been chosen to be tested on due to their unique properties. Pyrollidine was chosen because it is a similar, yet less complex heterocycle with nitrogen embedded in the ring. Cyclohexylamine was chosen to test the system's ability to degrade compounds that have the nitrogen as a substituent of a ring. This could allow us to independently test the functionality of genes which perform the 2nd half of carbazole degradation. Finally, 4-PBAH was chosen as it is a complex naphthenic acid that can serve as a model containing two of OSCAR's target compounds (Nitrogen and Carboxylic acid).
Why Use Synthetic Biology To Accomplish This Goal? Why Not Chemical Methods?
The most widely used chemical method for removing nitrogen from fuel sources is called hydrodenitrogenation (HDN). This process is not very efficient, as only 77% of nitrogen containing compounds are actually removed (Zeuthen et al, 2001). It also requires harsh conditions, for example temperatures upwards of 350 deg C and pressures up to 30 Bar. This is mostly because the nitrogen atoms in the ring must be hydrogenated before the carbon-nitrogen bond can be cleaved because this (Katzer & Sivasubramanian, 1979). The input of molybdenum based chemical catalysts required for this reaction is also very costly and can produce toxic by-products of its own (Zhu et al, 2008). While these chemical processes are a reasonable method of fuel upregulating they do have major limitations that are difficult to get around from a chemical standpoint. Synthetic biology can potentially offer a new avenue to address these problems, with much more potential for innovation and new ideas.
How Are We Going To Accomplish This?
The goal was to use enzymes produced by the Pseudomonas genus that are responsible for the conversion of carbazole to cathecol with the hope that they will also be able to act on similar nitrogen heterocycles. To make the pathway more efficient we have removed some enzymes from the native pathway that do not directly interact with nitrogen groups, replaced the native promoter with a TetR repressible promoter from the registry with stronger transcription, and explored using alternative enzymes to accomplish some of the critical steps.
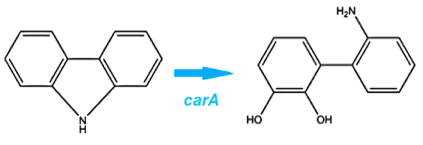
The first enzyme in this pathway is carbazole-1,9-dioxygenase (carA) which is responsible for selectively cleaving the first C-N bond in a nitrogen containing ring structure (Xu et al, 2006). This is a 4 part enzyme coded by 3 genes (2x carAa, carAc, and carAd). It converts carbazole into 2'-aminobiphenyl-2,3-diol. (Morales, 2010)
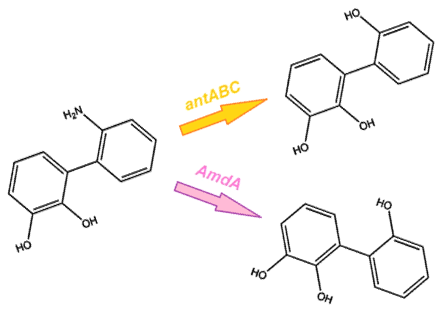
After this step the nitrogen is broken from the ring, however it is still attached to the structure. From this point, there are two options for the removal of nitrogen from the structure. The first option is the anthranilate 1,2-dioxygenase enzyme, coded by the ant operon in the native carbazole degradation pathway used by Pseudomonas (Diaz, 2010). While this option may seem the most reliable method, it does come with some limitations. It is designed to act on the substrate anthranilate, which is produced in the native pathway by the actions of the carB and carC enzymes, however since our synthetic pathway does not include these genes as they do not directly attack nitrogen there is a chance that this enzyme will not interact with our remaining nitrogen group. To this end, we have also biobricked a deaminase enzyme from Rhodococcus erythropolis that has been shown to selectively cleave the 2nd C-N bond from a variety of nitrogen heterocycles (Kilbane et al., 2002) (Kayser & Kilbane, 2005). This allows an alternative pathway for the 2nd step of nitrogen removal, possibly making the system more efficient and should show less substrate specificity than the ant genes allowing us to degrade a wider range of nitrogen containing rings.
Our Constructs
The carA genes have been biobricked individually into an operon under the control of a TetR promoter (BBa_J13002), with ribosome binding sites (BBa_B0034) inserted in front of each gene. The carAd gene was given a silent mutation to eliminate an illegal NotI site in the native sequence. The ant genes were biobricked as a whole operon with native ribosome binding sites in between each gene, also downstream of a TetR promoter. The AmdA gene was also biobricked downstream of a TetR promoter and ribosome binding site.
Using these constructs above 3 test circuits can be built. The carA operon is necessary for all three since it is the only way of accomplishing the first step in the pathway, however the AmdA and ant genes can both be used for the second step. This allows the construction of circuits which can independently test the ability of both systems to perform the final step of the pathway, as well as testing them together to see if having two pathways available makes the system significantly more efficient.
What Have We Shown So Far?
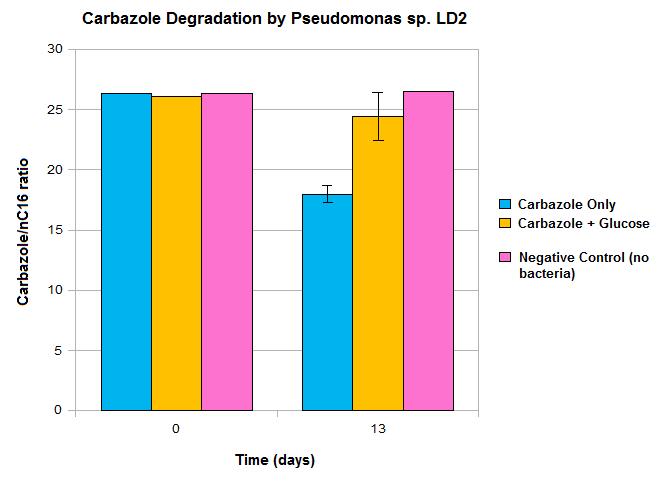
We have demonstrated the ability of Pseudomonas sp. LD2 to degrade carbazole, confirming that the genes we are biobricking do actually perform the desired function. However, these results also show some limitations in the native pathway that we hope our synthetic construct can address. Firstly, we only see modest loss of carbazole after 13 days, which would not be quick enough for OSCAR's large scale bioreactor. Secondly, the system must be glucose free for any significant carbazole loss to occur, which may not be realistic in our bioreactor. This may be due to the Pant promoter becoming deactivated in the presence of a preferred fuel source such as glucose and can be addressed by the introduction of a synthetic promoter. This promoter should also upregulate transcription levels, making the degradation process quicker. The alternative AmdA pathway should also allow the system to perform carbazole degradation more efficiently than we see in the native organism.
 "
"