Team:University College London/LabBook/Week15
From 2012.igem.org
Sednanalien (Talk | contribs) (→Tuesday (18.09.12)) |
Sednanalien (Talk | contribs) (→15-7) |
||
| Line 427: | Line 427: | ||
<div class="experimentContent"></html> | <div class="experimentContent"></html> | ||
| - | == | + | == Friday (21.09.12) == |
| + | |||
| + | |||
| + | |||
| + | '''Aim''' – We would like to determine if our laccase producing BBa_K729006 is capable of degrading polyethylene. | ||
| + | |||
| + | |||
| + | |||
<html></div> | <html></div> | ||
| Line 433: | Line 440: | ||
<div class="experimentContent"></html> | <div class="experimentContent"></html> | ||
| + | |||
==15-8== | ==15-8== | ||
Revision as of 01:40, 27 September 2012



Contents |
15.1 Monday 17.09.12
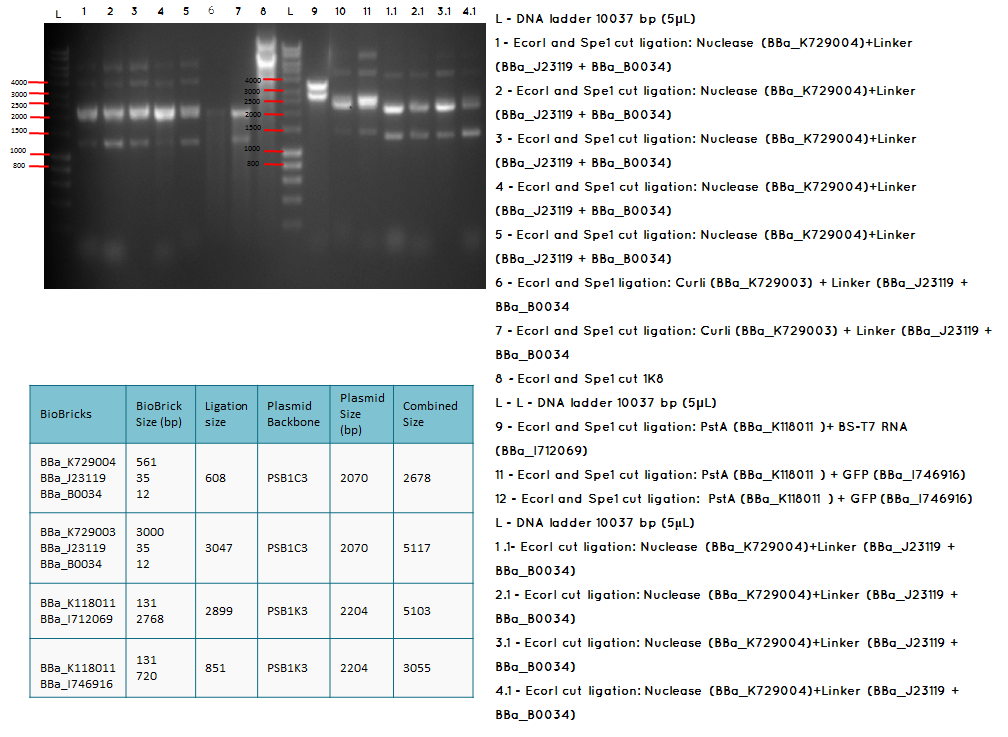
Aim - We want to check if the ligations have been successfulccessful
Step 1 We carried analytical digest of the following ligations:
- BBa_K729004 + BBa_J23119-BBa_B0034
- BBa_K729003 + BBa_J23119-BBa_B0034
- BBa_K118011 + BBA_I746916
Methods
Step 1 - Thawing cells: Thaw all materials on ice
Step 2 - Adding Ingredient: Add the following ingredients to autoclaved/sterile eppendorf tubes
| Component | Amount (ul) (one enzyme used) | Amount (ul) (two enzymes used) |
|---|---|---|
| dH20 | 2.5 | 1.5 |
| Buffer 1x | 1 | 1 |
| DNA template | 5 | 5 |
| BSA | 0.5 | 0.5 |
| Enzyme 1 | 1 | 2 |
| Enzyme 2 | N/A | 1 |
Step 3 - Addition of BioBrick: Flick contents gently and centrifuge.
Step 4 - Centrifuge:
RPM: 14000
Time: 1 minute
Temperature: 18oC
Step 5 - Digest Program: Place the samples on a thermocycler under the following conditions:
RPM: 550
Time: 2.5 hours
Temperature: 37oC
Step 6 - Denaturing Enzymes: If you are not running the samples on a gel immediately, denature the restriction enzymes by running the samples on a thermocycler under the following conditions:
RPM: 550
Time: 25 minutes
Temperature: 65oC
Step 2- We run a gel of the digests
Methods:
Preparing the Gel
Step 1: Within a conical flask, add 3ml 50X TAE, 1.5g Agarose, and 150ml RO water
Step 2: Heat for 1 min in microwave. Swirl. Heat again for 30s. If solution is clear stop. Else repeat.
Step 3: Cool solution under running cold water.
Step 4: Add 20ul ethidium bromide (normal concentration of EB solution is 500ug/ul)
Step 5: Pour into a sealed casting tray in a slow steady stream, ensuring there are no bubbles
Running a gel
Step 6: Add 1 part loading buffer to five parts of loading sample
Step 7: Position the gel in the tank and add TAE buffer, enough to cover the gel by several mm
Step 8: Add 5ul of DNA ladder to lane 1
Step 9: Add samples to the remaining wells
Step 10: Run at 100 volts for 1hour and 15 minutes
Imaging the Gel
Step 11: Place gel in GelDoc 2000 chamber
Step 12: Turn GelDoc 2000 chamber on
Step 13: From computer: Quantity One > Scanner > Gel_Doc_Xr>Manuqal Acquire
Step 14: Alter the exposure/settings to give a clear image.
TAE - Tris-acetate-EDTA
EDTA - ethylenediamine tetraacetic acid
Results:
Conclusion: The bands that we obtained on the gel did not correspond to the sizes we were expecting. For most of the samples we could clearly see a band at around 2000 which most probably is the backbone. These confusing results may be due to mistakes during the ligation or the enzyme digestion. Contamination is another possible reason for these results.

15.2
Tuesday (18.09.12)
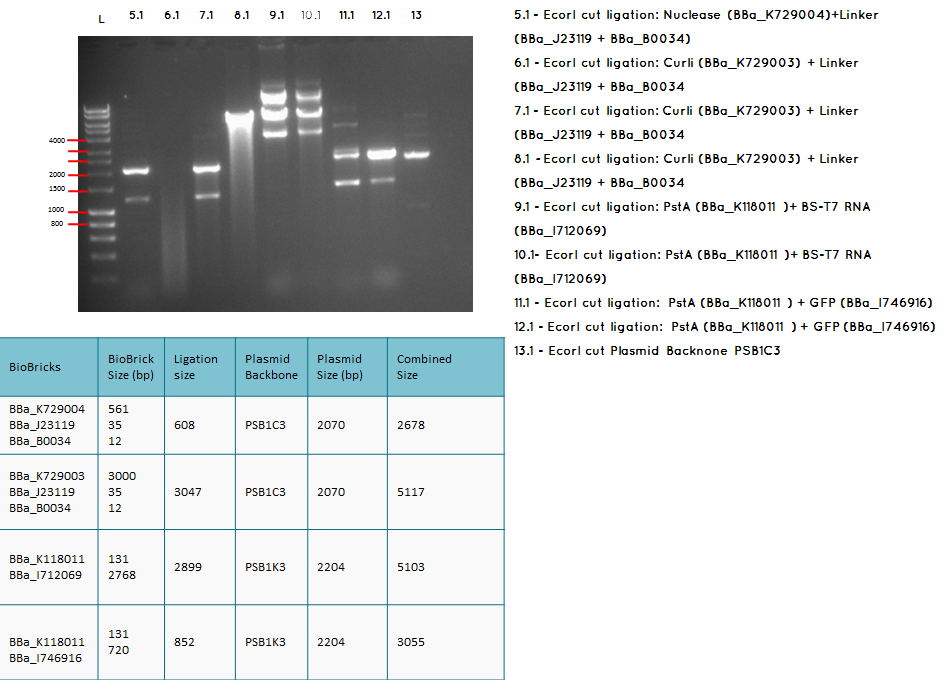
Aim - We want to check if the ligations have been successfulccessful
Step 1 We carried analytical digest of the following ligations:
- BBa_K729003+(BBa_J23119-BBa_B0034)
- BBa_K118011+BBa_I712069
- BBa_K729005+BBa_I712069
- BBa_I746909+K729007
Methods
Step 1 - Thawing cells: Thaw all materials on ice
Step 2 - Adding Ingredient: Add the following ingredients to autoclaved/sterile eppendorf tubes
| Component | Amount (ul) (one enzyme used) | Amount (ul) (two enzymes used) |
|---|---|---|
| dH20 | 2.5 | 1.5 |
| Buffer 1x | 1 | 1 |
| DNA template | 5 | 5 |
| BSA | 0.5 | 0.5 |
| Enzyme 1 | 1 | 2 |
| Enzyme 2 | N/A | 1 |
Step 3 - Addition of BioBrick: Flick contents gently and centrifuge.
Step 4 - Centrifuge:
RPM: 14000
Time: 1 minute
Temperature: 18oC
Step 5 - Digest Program: Place the samples on a thermocycler under the following conditions:
RPM: 550
Time: 2.5 hours
Temperature: 37oC
Step 6 - Denaturing Enzymes: If you are not running the samples on a gel immediately, denature the restriction enzymes by running the samples on a thermocycler under the following conditions:
RPM: 550
Time: 25 minutes
Temperature: 65oC
Step 2- We run a gel of the digests
Methods:
Preparing the Gel
Step 1: Within a conical flask, add 3ml 50X TAE, 1.5g Agarose, and 150ml RO water
Step 2: Heat for 1 min in microwave. Swirl. Heat again for 30s. If solution is clear stop. Else repeat.
Step 3: Cool solution under running cold water.
Step 4: Add 20ul ethidium bromide (normal concentration of EB solution is 500ug/ul)
Step 5: Pour into a sealed casting tray in a slow steady stream, ensuring there are no bubbles
Running a gel
Step 6: Add 1 part loading buffer to five parts of loading sample
Step 7: Position the gel in the tank and add TAE buffer, enough to cover the gel by several mm
Step 8: Add 5ul of DNA ladder to lane 1
Step 9: Add samples to the remaining wells
Step 10: Run at 100 volts for 1hour and 15 minutes
Imaging the Gel
Step 11: Place gel in GelDoc 2000 chamber
Step 12: Turn GelDoc 2000 chamber on
Step 13: From computer: Quantity One > Scanner > Gel_Doc_Xr>Manuqal Acquire
Step 14: Alter the exposure/settings to give a clear image.
TAE - Tris-acetate-EDTA
EDTA - ethylenediamine tetraacetic acid
Conclusion: We have obtained the expected sizes on for the pcstA (BBa_K118011) +RBS-T7RNAP (BBa_I712069) ligation.

Step 2 - Inoculating Colonies into a Selective Broth:: Add Yul of antibiotic to reach desired antibiotic concentration.
(For Ampicillin this is 50ug/ml, For Kanamycin it is 25ug/ml, for Tetracycline it is 15ug/ml, and for Chloramphenicol it is 25ug/ml)
Step 4 – Selecting a Colony: Select a clear, isolated colony and using an inoculation hoop scoop up a colony onto the tip. Deposit in the falcon tube
Step 5 - Culture: Culture your falcon tubes overnight at a temperature of 37 oC. Leave for no longer than 16 hours.
15.3 Tuesday 18.09.12
Aim – We would like to ligate BBa_I746909 to BBa_K729007 to obtain the following construct pCstA+RBS+T7RNAP+pT7+RBS+GFP+TT
Step 1 We minipreped the successful ligation of the pcstA (BBa_K118011) +RBS-T7RNAP (BBa_I712069) ligation which is our biobrick BBa_K729007
Method
Step 2 - Resuspend Cells: Resuspend pelleted bacterial cells in 250ul Buffer P1 and transfer to a microcentrifuge tube
Step 3 - Puncturing Cell Membrane: Add 250ul Buffer P2 and mix thoroughly by inverting the tube 4-6 times until the solution becomes clear. Do not allow the lysis reaction to proceed for more than 5 min.
Step 4 - Neutralising buffer P2: Add 350ul Buffer N3 and mix immediately and thoroughly by inverting the tube 4-6 times.
Step 5 - Centrifuge:
RPM: 13000
Time:10 minutes
Temperature: 18oC
Step 6 - Centrifuge: Apply the supernatant from step 5 to the QIAprep spin column by decanting or pipetting. Centrifuge for 30-60s and discard the flow-through.
Step 7 - Remove Endonucleases from Sample: Wash the QIAprep spin column by adding 500ul of Buffer PB. Centrifuge for 30-60s and discard flow-through.
Step 8 - Remove salts from sample: Wash the QIAprep spin column by adding 750ul of Buffer PE. Centrifuge for 3-60s and discard flow through.
Step 9 - Centrifuge:
RPM: 13000
Time:1 minute
Temperature: 18oC
Step 10 - Elute DNA: Place the QIAprep column in a clean 1.5ml microcentrifuge tube. To elute DNA, add 50ul Buffer EB to the centre of the spin column, let it stand for 1 min, and centrifuge for 1 min.
Step 2 Ligation was performed using the Biobricks BBa_I746909 and BBa_K729007 to generate a Biobrick that contains pcstA-T7RNAP-pT7-GFP-TT.
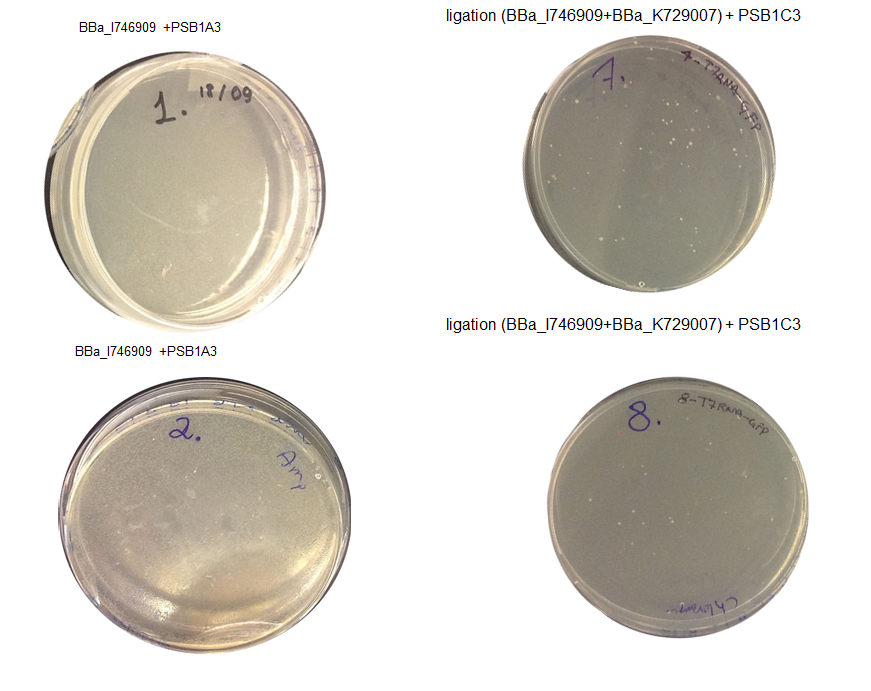
Step 3 Transformation – we plated 4 plates; plate 1 and 2 were with the transformed BBa_I746909 biobrick and plate 3 and 4 were transformed with our ligation (BBa_I746909+BBa_K729007). We are going to incubate over night at 37˚C and colonies picking the next day.
Method:
Step 1 - Addition of BioBrick: To the still frozen competent cells, add 1 - 5 µL of the resuspended DNA to the 2ml tube.
Step 4 - Incubation: Close tube and incubate the cells on ice for 45 minutes.
Step 5 - Heat Shock: Heat shock the cells by immersion in a pre-heated water bath at 37ºC for 10 minutes.
Step 6 - Incubation: Incubate the cells on ice for 2 minutes.
Step 7 - Add media: Add 1.5ml of Lysogeny Broth and transfer to a falcon tube.
Step 8 - Incubation: Incubate the cells at 37ºC for 1 hour at RPM 550.
Step 9 - Transfer: transfer the solution back into a 1.5ml Eppendorf and centrifuge
RPM: 14000
Time: 2 minutes
Temperature (18-25oC)
Step 10 - Resuspend:Remove supernatant and resuspend in 100ul LB
Step 11 - Plating: Spread the resuspended cell solution onto a selective nutrient agar plate. Place the plates in a 37°C static incubator, leave overnight (alternatively a 30°C static incubator over the weekend)
Aim 1 – Check the result of transformation. We expect to see growth on plates with transformed biobricks and ligations.
Results - The table below indicates that there was growth on all of the plates
| Plate | Samples | Volume Inoculated | Colony Formation |
|---|---|---|---|
| 1 | BioBrick BBa_I746909 | 10ul | Yes |
| 2 | BioBrick BBa_I746909 | 90uL | Yes |
| 3 | Ligation BBa_I746909+BBa_K729007 | 10ul | Yes |
| 4 | Ligation BBa_I746909+BBa_K729007 | 90uL | Yes |
Conclusion : The transformations have been successful. Colonies will be picked and inoculated into LB media for overnight culture

Tuesday (18.09.12)
Aim – We would like to ligate BBa_I746909 to BBa_K729007 to obtain the following construct pCstA+RBS+T7RNAP+pT7+RBS+GFP+TT
Step 1 We minipreped the successful ligation of the pcstA (BBa_K118011) +RBS-T7RNAP (BBa_I712069) ligation which is our BioBrick BBa_K729007
Method
Step 2 - Resuspend Cells: Resuspend pelleted bacterial cells in 250ul Buffer P1 and transfer to a microcentrifuge tube
Step 3 - Puncturing Cell Membrane: Add 250ul Buffer P2 and mix thoroughly by inverting the tube 4-6 times until the solution becomes clear. Do not allow the lysis reaction to proceed for more than 5 min.
Step 4 - Neutralising buffer P2: Add 350ul Buffer N3 and mix immediately and thoroughly by inverting the tube 4-6 times.
Step 5 - Centrifuge:
RPM: 13000
Time:10 minutes
Temperature: 18oC
Step 6 - Centrifuge: Apply the supernatant from step 5 to the QIAprep spin column by decanting or pipetting. Centrifuge for 30-60s and discard the flow-through.
Step 7 - Remove Endonucleases from Sample: Wash the QIAprep spin column by adding 500ul of Buffer PB. Centrifuge for 30-60s and discard flow-through.
Step 8 - Remove salts from sample: Wash the QIAprep spin column by adding 750ul of Buffer PE. Centrifuge for 3-60s and discard flow through.
Step 9 - Centrifuge:
RPM: 13000
Time:1 minute
Temperature: 18oC
Step 10 - Elute DNA: Place the QIAprep column in a clean 1.5ml microcentrifuge tube. To elute DNA, add 50ul Buffer EB to the centre of the spin column, let it stand for 1 min, and centrifuge for 1 min.
Step 2 Ligation was performed using the BioBricks BBa_I746909 and BBa_K729007 to generate a BioBrick that contains pcstA-T7RNAP-pT7-GFP-TT.
Step 3 Transformation – we plated 4 plates; plate 1 and 2 were with the transformed BBa_I746909 biobrick and plate 3 and 4 were transformed with our ligation (BBa_I746909+BBa_K729007). We are going to incubate over night at 37C and colonies picking the next day.
Method:
Step 1 - Addition of BioBrick: To the still frozen competent cells, add 1 - 5 µL of the resuspended DNA to the 2ml tube.
Step 4 - Incubation: Close tube and incubate the cells on ice for 45 minutes.
Step 5 - Heat Shock: Heat shock the cells by immersion in a pre-heated water bath at 37ºC for 10 minutes.
Step 6 - Incubation: Incubate the cells on ice for 2 minutes.
Step 7 - Add media: Add 1.5ml of Lysogeny Broth and transfer to a falcon tube.
Step 8 - Incubation: Incubate the cells at 37ºC for 1 hour at RPM 550.
Step 9 - Transfer: transfer the solution back into a 1.5ml Eppendorf and centrifuge
RPM: 14000
Time: 2 minutes
Temperature (18-25oC)
Step 10 - Resuspend:Remove supernatant and resuspend in 100ul LB
Step 11 - Plating: Spread the resuspended cell solution onto a selective nutrient agar plate. Place the plates in a 37°C static incubator, leave overnight (alternatively a 30°C static incubator over the weekend)
Wednesday (19.09.12)
Aim 1 – Check the result of transformation. We expect to see growth on plates with transformed biobricks and ligations.
Results - The table below indicates that there was growth on all of the plates
| Plate | Samples | Volume Inoculated | Colony Formation |
|---|---|---|---|
| 1 | BioBrick BBa_I746909 | 10ul | Yes |
| 2 | BioBrick BBa_I746909 | 90uL | Yes |
| 3 | Ligation BBa_I746909+BBa_K729007 | 10ul | Yes |
| 4 | Ligation BBa_I746909+BBa_K729007 | 90uL | Yes |
Conclusion : The transformations have been successful. Colonies will be picked and inoculated into LB media for overnight culture
Step 4 – Inoculating Colonies into a Selective Broth. We had colonies on all the plates. We picked we picked 2 colonies from plates 1 and 2 and 4 colonies from plate 3 and 4. We grow all 12 colonies overnight.The table below indicates the volume of broth and the concentration of antibiotic required.
Step 2 - Inoculating Colonies into a Selective Broth:: Add Yul of antibiotic to reach desired antibiotic concentration.
(For Ampicillin this is 50ug/ml, For Kanamycin it is 25ug/ml, for Tetracycline it is 15ug/ml, and for Chloramphenicol it is 25ug/ml)
Step 4 – Selecting a Colony: Select a clear, isolated colony and using an inoculation hoop scoop up a colony onto the tip. Deposit in the falcon tube
Step 5 - Culture: Culture your falcon tubes overnight at a temperature of 37 oC. Leave for no longer than 16 hours.
| Falcon tube | Samples | Broth | Antibiotic (50 nu/uL) |
|---|---|---|---|
| 1 | BioBrick BBa_I746909 | Lysogeny Broth | Ampicillinn |
| 2 | BioBrick BBa_I746909 | Lysogeny Broth | Ampicillin |
| 3 | BioBrick BBa_I746909 | Lysogeny Broth | Ampicillin |
| 4 | BioBrick BBa_I746909 | Lysogeny Broth | Ampicillin |
| 5 | Ligation BBa_I746909+BBa_K729007 | Lysogeny Broth | Chloramphenicol |
| 6 | Ligation BBa_I746909+BBa_K729007 | Lysogeny Broth | Chloramphenicol |
| 7 | Ligation BBa_I746909+BBa_K729007 | Lysogeny Broth | Chloramphenicol |
| 8 | Ligation BBa_I746909+BBa_K729007 | Lysogeny Broth | Chloramphenicol |
| 9 | Ligation BBa_I746909+BBa_K729007 | Lysogeny Broth | Chloramphenicol |
| 10 | Ligation BBa_I746909+BBa_K729007 | Lysogeny Broth | Chloramphenicol |
| 11 | Ligation BBa_I746909+BBa_K729007 | Lysogeny Broth | Chloramphenicol |
| 12 | Ligation BBa_I746909+BBa_K729007 | Lysogeny Broth | Chloramphenicol |
Thursday (20.09.12)
Aim - We want to check if the ligations were successful.
All 12 colonies so we are going to miniprep and nanodrop all of them
Step 1 - Miniprep
Method
Step 2 - Resuspend Cells: Resuspend pelleted bacterial cells in 250ul Buffer P1 and transfer to a microcentrifuge tube
Step 3 - Puncturing Cell Membrane: Add 250ul Buffer P2 and mix thoroughly by inverting the tube 4-6 times until the solution becomes clear. Do not allow the lysis reaction to proceed for more than 5 min.
Step 4 - Neutralising buffer P2: Add 350ul Buffer N3 and mix immediately and thoroughly by inverting the tube 4-6 times.
Step 5 - Centrifuge:
RPM: 13000
Time:10 minutes
Temperature: 18oC
Step 6 - Centrifuge: Apply the supernatant from step 5 to the QIAprep spin column by decanting or pipetting. Centrifuge for 30-60s and discard the flow-through.
Step 7 - Remove Endonucleases from Sample: Wash the QIAprep spin column by adding 500ul of Buffer PB. Centrifuge for 30-60s and discard flow-through.
Step 8 - Remove salts from sample: Wash the QIAprep spin column by adding 750ul of Buffer PE. Centrifuge for 3-60s and discard flow through.
Step 9 - Centrifuge:
RPM: 13000
Time:1 minute
Temperature: 18oC
Step 10 - Elute DNA: Place the QIAprep column in a clean 1.5ml microcentrifuge tube. To elute DNA, add 50ul Buffer EB to the centre of the spin column, let it stand for 1 min, and centrifuge for 1 min.
Step 2 - Nanodrop
Method
Software ND-1000 Model:
Step 1: Initialise the spectrophotometer by pipetting 1 µ of clean water onto lower optic surface, lowering the lever arm and selecting ‘initialise’ in the ND-1000 software
Step 2: Wipe and add elution buffer as negative control. Click blank in ND-1000 software
Step 3: Wipe and add 1 µl sample
Step 4: On the software set lambda to 260nm
Step 5: Lower the lever arm and click measure in ND-1000 software
Step 6: Take readings for concentration and purity
Step 7: Once measurement complete, wipe surface
In the table below are summarised all the concentrations obtained from the nanodrop.
| Falcon tube | Samples | Plate | Concentration at λ260 (ng/uL) |
|---|---|---|---|
| 1 | BioBrick BBa_I746909 | 1 | 188 |
| 2 | BioBrick BBa_I746909 | 1 | 276.2 |
| 3 | BioBrick BBa_I746909 | 2 | 194.6 |
| 4 | BioBrick BBa_I746909 | 2 | 329.6 |
| 5 | Ligation BBa_I746909+BBa_K729007 | 3 | 233.6 |
| 6 | Ligation BBa_I746909+BBa_K729007 | 3 | 197.7 |
| 7 | Ligation BBa_I746909+BBa_K729007 | 3 | 438.5 |
| 8 | Ligation BBa_I746909+BBa_K729007 | 3 | 337.8 |
| 9 | Ligation BBa_I746909+BBa_K729007 | 4 | 310.3 |
| 10 | Ligation BBa_I746909+BBa_K729007 | 4 | 244.3 |
| 11 | Ligation BBa_I746909+BBa_K729007 | 4 | 249.8 |
| 12 | Ligation BBa_I746909+BBa_K729007 | 4 | 340.2 |
Step 3 - We carried out analytical digest of the minipreped DNA
Methods
Step 1 - Thawing cells: Thaw all materials on ice
Step 2 - Adding Ingredient: Add the following ingredients to autoclaved/sterile eppendorf tubes
| Component | Amount (ul) (one enzyme used) | Amount (ul) (two enzymes used) |
|---|---|---|
| dH20 | 2.5 | 1.5 |
| Buffer 1x | 1 | 1 |
| DNA template | 5 | 5 |
| BSA | 0.5 | 0.5 |
| Enzyme 1 | 1 | 2 |
| Enzyme 2 | N/A | 1 |
Step 3 - Addition of BioBrick: Flick contents gently and centrifuge.
Step 4 - Centrifuge:
RPM: 14000
Time: 1 minute
Temperature: 18oC
Step 5 - Digest Program: Place the samples on a thermocycler under the following conditions:
RPM: 550
Time: 2.5 hours
Temperature: 37oC
Step 6 - Denaturing Enzymes: If you are not running the samples on a gel immediately, denature the restriction enzymes by running the samples on a thermocycler under the following conditions:
RPM: 550
Time: 25 minutes
Temperature: 65oC
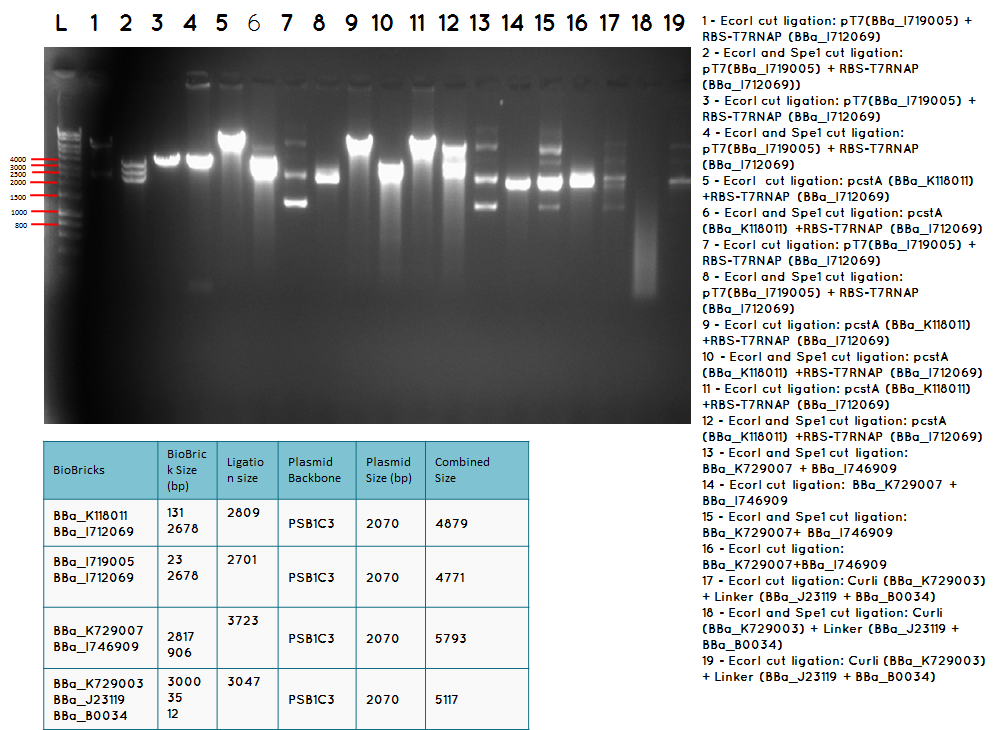
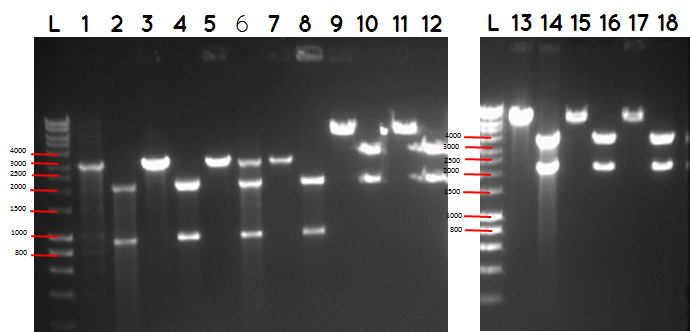
step 4 - Gel electrophoresis of the analytical digest to see if we have the right DNA size
L - DNA ladder 10037 bp (5μL)
1 - PstI cut BBa_I746909+PSB1A3
2 - PstI and Xbal cut BBa_I746909+PSB1A3
3 - PstI cut BBa_I746909+PSB1A3
4 - PstI and Xbal cut BBa_I746909+PSB1A3
5 - PstI cut BBa_I746909+PSB1A3
6 - PstI and Xbal cut BBa_I746909+PSB1A3
7 - PstI cut BBa_I746909+PSB1A3
8 - PstI and Xbal cut BBa_I746909+PSB1A3
9 – PstI cut ligation (BBa_K729007+BBa_I746909) + PSB1C3
10 - PstI and Xbal cut ligation (BBa_K729007+BBa_I746909) + PSB1C3
11 - PstI cut ligation (BBa_K729007+BBa_I746909) + PSB1C3
12 - PstI and Xbal cut ligation (BBa_K729007+BBa_I746909) + PSB1C3
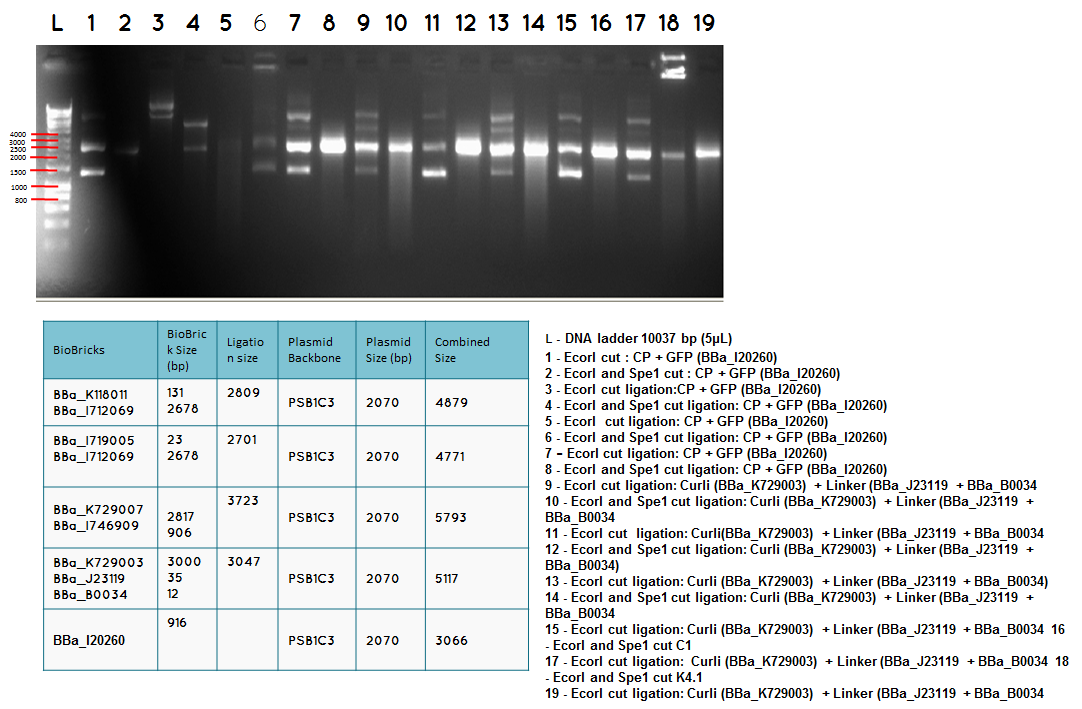
L DNA ladder 10037 bp (5μL)
13 - PstI cut ligation (BBa_K729007+BBa_I746909) + PSB1C3
14 - PstI and Xbal cut ligation (BBa_K729007+BBa_I746909) + PSB1C3
15 - PstI cut ligation (BBa_K729007+BBa_I746909) + PSB1C3
16 - PstI and Xbal cut ligation (BBa_K729007+BBa_I746909) + PSB1C3
17 - PstI cut ligation (BBa_K729007+BBa_I746909) + PSB1C3
18 - PstI and Xbal cut ligation (BBa_K729007+BBa_I746909) + PSB1C3
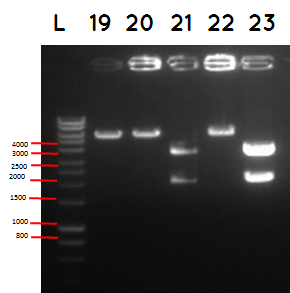
L - DNA ladder 10037 bp (5μL)
19 - PstI cut ligation (BBa_K729007+BBa_I746909) + pSB1C3
20 - PstI cut cut ligation (BBa_K729007+BBa_I746909) + pSB1C3
21 - PstI and Xbal cut ligation (BBa_K729007+BBa_I746909) + pSB1C3
22 - PstI cut ligation (BBa_K729007+BBa_I746909) + pSB1C3
23 -PstI and Xbal cut ligation (BBa_K729007+BBa_I746909) + pSB1C3
| BioBricks | BioBrick size (bp) | Ligation size (bp) | Plasmid backbone | Pasmid backbone(bp) | Combined size(bp) |
|---|---|---|---|---|---|
| BBa_K729007+BBa_I746909 | 2817+906 | 3731 | pSB1C3 | 2070 | 5801 |
| BBa_I746909 | 906 | pSB1A3 | 2155 | 3061 |
Conclusion - The ligation was successful the bands on the gel correspond to the bands we were expecting.

15-5

Tuesday (18.09.12)
Aim – We would like to determine if BBa_K729006 is functioning as intended and producing laccase.
1) Cells are grown overnight in 10ml of LB media at 37˚C, 200rpm.
- a) BBa_K729006
- b) W3110 Control
2) Cells are centrifuged at 6100g for 20 minutes, collecting the supernatant containing laccase.
3) Cuvettes are filled with 2.2ml KH2PO4 buffer, 0.5 ml laccase supernatant, and 0.3 ml Syringaldazine. Blank replaces the supernatant with LB media.
4) Optical density at 530nm is measured at 5 minute intervals for 30 minutes.
Conclusion - Our cells are producing an elevate amount of laccase compared to the control, indicating that the BioBrick is functioning as intended.

Friday (21.09.12)
Aim – We would like to determine if our laccase producing BBa_K729006 is capable of degrading polyethylene.

 "
"