Team:Calgary/Notebook/Hydrocarbon
From 2012.igem.org
| (45 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{Team:Calgary/MainHeader}} | ||
| + | {{Team:Calgary/BasicPage| | ||
| + | TITLE=Team| | ||
| + | CONTENT= | ||
<html> | <html> | ||
<!-- | <!-- | ||
| Line 6: | Line 10: | ||
--> | --> | ||
| - | < | + | <i><h3 style="font-family: Arial;">THIS PAGE NO LONGER EXISTS AND WILL NOT APPEAR ON THE WIKI. Please migrate your data and information to your respective pages (as outlined in the email I sent you a while back.) -PW</h3></i> |
| + | |||
| + | <h3>you guys need to update your notebook more?</h3> | ||
| + | <h3>we working hard, y u get so mad? :'(</h3> | ||
<h2>Week 1 (May 1-4)</h2> | <h2>Week 1 (May 1-4)</h2> | ||
<p>The following section covers wetlab aspect of our overall project focusing on microbial conversion of naphthenic acids into economically-valuable hydrocarbons. The approach taken in this endeavour will be from four strategic starting points - ring cleavage, decarboxylation, denitrificaiton, and desulfurization. Overall, the 'hydrocarbons' aspect of the project is a critical one to our overall design and construction of a biosystem capable of not only detecting but also converting naphthenic acids in what will be an economically viable solution to the remediation and recovery of tailings pond water, while removing these toxic compounds before they can have significant detrimental impact on the environment. </p> | <p>The following section covers wetlab aspect of our overall project focusing on microbial conversion of naphthenic acids into economically-valuable hydrocarbons. The approach taken in this endeavour will be from four strategic starting points - ring cleavage, decarboxylation, denitrificaiton, and desulfurization. Overall, the 'hydrocarbons' aspect of the project is a critical one to our overall design and construction of a biosystem capable of not only detecting but also converting naphthenic acids in what will be an economically viable solution to the remediation and recovery of tailings pond water, while removing these toxic compounds before they can have significant detrimental impact on the environment. </p> | ||
| + | |||
<h2>Week 2 (May 7-11)</h2> | <h2>Week 2 (May 7-11)</h2> | ||
<h3>Decarboxylation</h3> | <h3>Decarboxylation</h3> | ||
| Line 18: | Line 26: | ||
<p>In addition to our research we have also been learning some of the lab techniques we will be using this summer. This includes transforming a plasmid into E. coli, plating and selecting for bacteria containing the plasmid, verifying with colony PCR, performing a mini-prep and a restriction digest.</p> | <p>In addition to our research we have also been learning some of the lab techniques we will be using this summer. This includes transforming a plasmid into E. coli, plating and selecting for bacteria containing the plasmid, verifying with colony PCR, performing a mini-prep and a restriction digest.</p> | ||
<h3>Desulfurization</h3> | <h3>Desulfurization</h3> | ||
| - | <p> | + | <p>Along with the rest of the team, this week was dedicated to familiarizing ourselves on the protocols that will be utilized during this years project; specifically the polymerase chain reaction, gel verification, preparation of overnight cultures, as well as developing a procedural flowchart to transform competent cells with registry biobricks. With regards to our sub-group specific goals, we reviewed the current available literature around various industrial and laboratory approaches to desulfurization of organic groups, especially in the petroleum industry. This included a comparison of non-biological processes such as conventional hydrodesulfurization, which is currently employed in petroleum product refinery stages, and how a biological approach would supplement and perhaps even offer several advantages over these methods. Current limitations to biological desulfurization, however, include such factors as biocatalyst stability, enzyme specificity, desulfurization rate, and a need for a carbon source to regenerate co-factors. We also identified the enzyme desulfinase (DszB) as being one of the bottlenecks in the desulfurization 4S pathway. Overall, our goals moving forward involve determining the specific pathways involved in the desulfurization process as well as the reaction conditions we would want to employ, and identifying specific model compounds in addition to dibenzothiophene (DBT) that we could use to test the effectivity of our biosystem in order to determine its functionality in the conversion of naphthenic acids to economically valuable hydrocarbons.</p> |
<h3>Ring Cleavage</h3> | <h3>Ring Cleavage</h3> | ||
<p>This week we mainly researched aromatic ring cleaving using intra- and extradiol dioxygenases from species of Pseudomonas and Bacillus but also began literature searches on aliphatic ring cleavage done by monooxygenases. </p> | <p>This week we mainly researched aromatic ring cleaving using intra- and extradiol dioxygenases from species of Pseudomonas and Bacillus but also began literature searches on aliphatic ring cleavage done by monooxygenases. </p> | ||
| + | |||
<h2>Week 3 (May 14-18)</h2> | <h2>Week 3 (May 14-18)</h2> | ||
<h3>Decarboxylation</h3> | <h3>Decarboxylation</h3> | ||
| Line 27: | Line 36: | ||
<p>In the second week we continued to do more research on the degradation of alicyclic compounds and found two strains of bacteria that contained genes needed for this process. The first strain, Thauera butanivorans, contains the genes required to activate the ring by adding a hydroxyl group. The gene is called butane monooxygenase and is composed of three subunits, a hydroxylase, a reductase, and a regulatory component. The second strain, Acinetobacter sp. SE19, contains the genes needed to oxidize the alcohol, formed by butane monooxygenase, and to cleave the ring. There is a cluster of nine genes that perform this oxidation and cleavage but only six are involved directly.</p> | <p>In the second week we continued to do more research on the degradation of alicyclic compounds and found two strains of bacteria that contained genes needed for this process. The first strain, Thauera butanivorans, contains the genes required to activate the ring by adding a hydroxyl group. The gene is called butane monooxygenase and is composed of three subunits, a hydroxylase, a reductase, and a regulatory component. The second strain, Acinetobacter sp. SE19, contains the genes needed to oxidize the alcohol, formed by butane monooxygenase, and to cleave the ring. There is a cluster of nine genes that perform this oxidation and cleavage but only six are involved directly.</p> | ||
<h3>Desulfurization</h3> | <h3>Desulfurization</h3> | ||
| - | <p> Building on the previous week's literature review, the 4S pathway was | + | <p> Building on the previous week's literature review, the 4S pathway was recognized as the preferred biological mechanism that we would explore in devising a desulfurization biosystem. Of specific interest is the dsz operon consisting of the genes for <i> dszA, dszB, </i>and<i> dszC</i> which selectively and non-destructively remove the sulfur from the hydrocarbon structure, and therefore preserves the carbon skeleton. In addition to these, another dsz gene exists.<i> dszD</i>, which codes for a FMN:NADH reductase, is an essential component of the pathway, but not part of the operon. Instead, it is encoded on the chromosome. The enzyme produced by this gene is required to regenerate the FMNH2 consumed by the reactions carried out by DszA and DszC. <i>Rhodococcus erythropolis</i> IGTS8 is the most studied model organism in investigations of the 4S pathway, and has been shown in many different research endeavors to be capable of converting DBT to 2-HBP.</p><p> |
| + | </p><b> INSERT PATHWAY DIAGRAM </b><p> | ||
| + | An alternative to the DszD gene is HpaC, an oxidoreductase encoded in the <i>E. coli</i> W genome. This enzyme has been shown to increase the rate of desulfurization by <b>x amount find citation </b> Following this, other protocols added to our growing lab methods 'toolkit' were a restriction digest protocol, PCR purification, and finally, DNA construction digest. Aims moving forward include obtaining strains of the <i> R. erythropolis </i>, while also executing a timeline devised to biobrick, test, and incorporate the genes necessary in the above processes in a biobrick circuit.</p> | ||
<h2>Week 4 (May 22-25)</h2> | <h2>Week 4 (May 22-25)</h2> | ||
| Line 55: | Line 66: | ||
<h3>Desulfurization</h3> | <h3>Desulfurization</h3> | ||
| - | <p>This week was kicked off with a project development meeting with Emily and David, and we devised a protocol for biobricking the | + | |
| - | Furthermore, we ordered the substrates/compounds that we | + | <p>This week was kicked off with a project development meeting with Emily and David, and we devised a protocol for biobricking the <i> hpaC </i> gene. Additionally, methods to place the genes coding for the 4 enzymes, DszA,B,C and HpaC into a single construct were explored. Within the lab, the PCR performed on the resuspended pUC18-hpaC was not successful initially. |
| + | Furthermore, we ordered the substrates/compounds that we intend to use for desulfurization tests. Once the substrates and the <i>Rhodococcus</i> strain arrive we are going to test how effectively the bacteria can desulfurize different sulphur-containing compounds that resemble naphthenic acids. Finally, we came across a paper published by <b> REFERENCE </b>, whos team had developed an improved efficiency DszB through site-directed mutagenesis in 2007. This was through a point mutation to the gene, converting a tyrosine at position 63 to a phenylalanine residue. A member of this team was contacted to request the plasmid that contains the mutated gene. The conversion step carried out by DszB is the major bottleneck in the 4S pathway and if a strain or sample containing this mutation was obtained, it would significantly bolster our later testing efforts on DBT, as well as other compounds such as thiophane. | ||
</p> | </p> | ||
| - | + | ||
<h3>Denitrification</h3> | <h3>Denitrification</h3> | ||
<p>This week we reviewed the primers listed in the database and also designed some new ones. Primers for CarAa, CarAc, and CarAd were designed individually, while primers for CarBaBbC and AntABC were designed to encompass multiple genes in a sequence. These primers were designed to be used on Pseudomonas putida which we decided to use as our gene source since it was available to us as opposed to ordering Pseudomonas resinovorans. We also designed a primer for the AmdA gene from Rhodococcus erthyroplois. In addition to ordering these primers we also ordered the nitrogen containing compounds that we will need to test these enzymes on. We decided on using carbazole to make sure the enzymes can perform their natural function as well as pyrrolidine to test them on a similar ring structure. We also ordered cyclohexamine in order to independently test the function of the alternative AmdA pathway. Finally, we decided to eventually order 4-Piperidine butyric acid hydrochloride to test how the enzymes will work on nitrogen containing naphthenic acids. However, we decided since it is a very expensive compound we would wait to make sure the enzyme's work on more simple compounds before ordering it.</p> | <p>This week we reviewed the primers listed in the database and also designed some new ones. Primers for CarAa, CarAc, and CarAd were designed individually, while primers for CarBaBbC and AntABC were designed to encompass multiple genes in a sequence. These primers were designed to be used on Pseudomonas putida which we decided to use as our gene source since it was available to us as opposed to ordering Pseudomonas resinovorans. We also designed a primer for the AmdA gene from Rhodococcus erthyroplois. In addition to ordering these primers we also ordered the nitrogen containing compounds that we will need to test these enzymes on. We decided on using carbazole to make sure the enzymes can perform their natural function as well as pyrrolidine to test them on a similar ring structure. We also ordered cyclohexamine in order to independently test the function of the alternative AmdA pathway. Finally, we decided to eventually order 4-Piperidine butyric acid hydrochloride to test how the enzymes will work on nitrogen containing naphthenic acids. However, we decided since it is a very expensive compound we would wait to make sure the enzyme's work on more simple compounds before ordering it.</p> | ||
| Line 83: | Line 95: | ||
<h3>Decarboxylation</h3> | <h3>Decarboxylation</h3> | ||
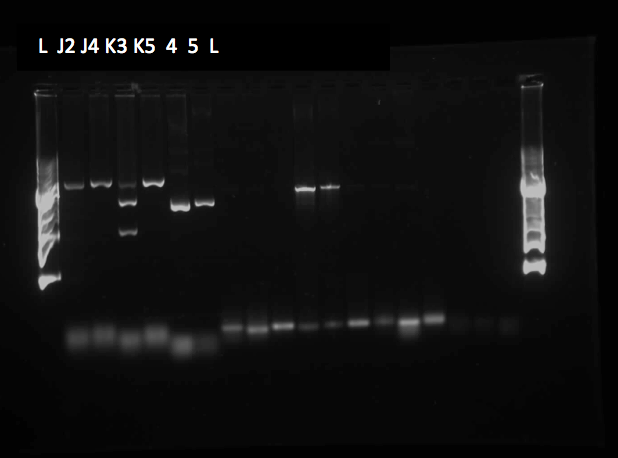
| - | <p> On Tuesday (May 29), a gel was run on the restriction digests of the extracted plasmids from Week 4, which appeared to confirm the successful transformation of the PetroBrick, showing clear bands at the pertinent locations. The gel is as follows: </p> | + | <p> On Tuesday (May 29), a gel was run on the restriction digests of the extracted plasmids from Week 4, which appeared to confirm the successful transformation of the PetroBrick, showing clear bands at the pertinent locations. The gel is as follows: </p> |
[[File:UCalgary igem PCRverfication Hydrocarbon PetroBrick Gel 2.PNG|thumb|500px|center]]<html> | [[File:UCalgary igem PCRverfication Hydrocarbon PetroBrick Gel 2.PNG|thumb|500px|center]]<html> | ||
| Line 108: | Line 120: | ||
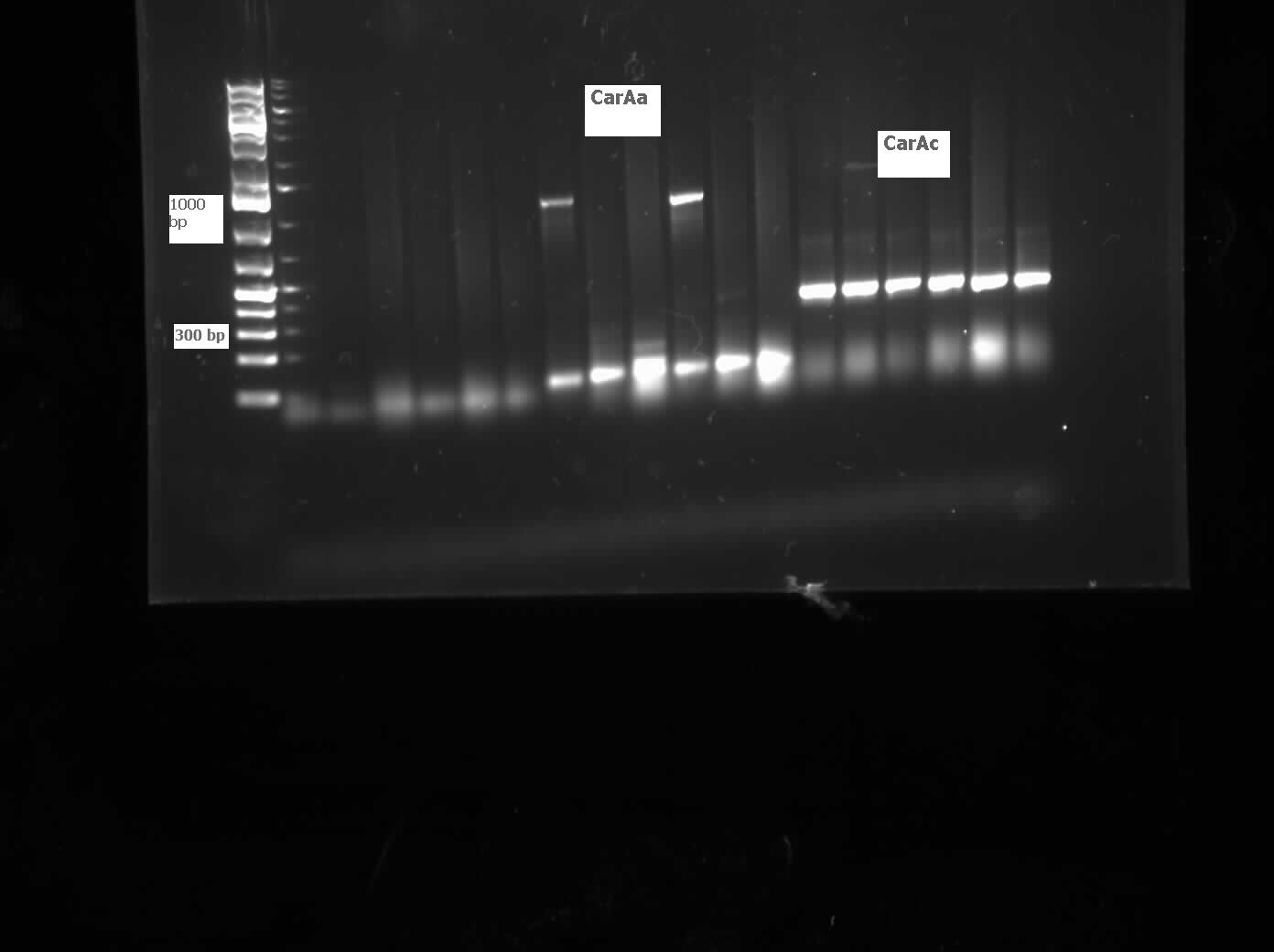
<p>On Monday and Tuesday we used the database primers to attempt to isolate CarAa, CarAc, CarAd, AntA, AntB, and AntC from the Psuedomonas putida we plated last week. CarAd, AntB, and AntC were all put in the gradient PCR machine to account for the wide range of their primers’ melting points, while the others were done via regular PCR. Unfortunately, no positive results were obtained from these reactions. The PCR on CarAc and AntA was repeated on Wednesday, still not giving positive results. Finally, we attempted to use a salt concentration gradient on the PCR reaction for CarAc and AntA, using concentrations that ranged from 1.0 microlitres/tube to 2.0 microlitres/tube in increments of 0.2 microlitres/tube. This helped as CarAc showed bands in samples that had concentrations of 1.2 and 1.4 microlitres/tube. AntA also showed bands, however they were not the correct size, indicating contamination and/or non-specific annealing of the primer. The positive control also showed bands of the correct size. Earlier in the week we also made overnight cultures of 3 environmental strains (28, 29, 30) of Psuedomonas putida from -80 glycerol stock. On Friday we performed a genomic prep on these cultures, and plan on attempting to amplify genes from the isolated DNA next week. </p> | <p>On Monday and Tuesday we used the database primers to attempt to isolate CarAa, CarAc, CarAd, AntA, AntB, and AntC from the Psuedomonas putida we plated last week. CarAd, AntB, and AntC were all put in the gradient PCR machine to account for the wide range of their primers’ melting points, while the others were done via regular PCR. Unfortunately, no positive results were obtained from these reactions. The PCR on CarAc and AntA was repeated on Wednesday, still not giving positive results. Finally, we attempted to use a salt concentration gradient on the PCR reaction for CarAc and AntA, using concentrations that ranged from 1.0 microlitres/tube to 2.0 microlitres/tube in increments of 0.2 microlitres/tube. This helped as CarAc showed bands in samples that had concentrations of 1.2 and 1.4 microlitres/tube. AntA also showed bands, however they were not the correct size, indicating contamination and/or non-specific annealing of the primer. The positive control also showed bands of the correct size. Earlier in the week we also made overnight cultures of 3 environmental strains (28, 29, 30) of Psuedomonas putida from -80 glycerol stock. On Friday we performed a genomic prep on these cultures, and plan on attempting to amplify genes from the isolated DNA next week. </p> | ||
<h3>Desulfurization</h3> | <h3>Desulfurization</h3> | ||
| - | <p>Since we wanted to make sure we would not run out of pUC18(plasmid containing hpaC gene), we transformed some E.coli cells with it. We grew them on plates containing A, K, T and C antibiotics and they only grew on A. Therefore pUC18 has A resistance. We did a three sets of PCR with | + | <p>Since we wanted to make sure we would not run out of pUC18(plasmid containing the hpaC gene), we transformed some E.coli cells with it. We grew them on plates containing A, K, T and C antibiotics and they only grew on A. Therefore pUC18 has A resistance. We did a three sets of PCR with primers designed against <i>hpaC</i>, one using 1/10 dilution of pUC18, the other using 1/100 dilution of pUC18 and one with the colonies we had just obtained by transforming the E.coli cells. The PCR worked and we saw bands of the same size for all three sets of PCR. (Unfortunately, the picture we saved is not a good one since some of the bands faded away under UV due to prolonged exposure. Following this, PCR purification was performed to obtain the pure <i>hpaC</i> with biobrick prefix and suffix attatched to gene, which would allow us to insert the sequence into a biobrick standard backbone. 3 sets of digestion, ligation, and transformation (using pairs of X&P enzymes, E&S enzymes and E&P enzymes) were carried out in order to insert the <i>hpaC </i> gene into the pSB1C3 vector. All the sets grew successfully. |
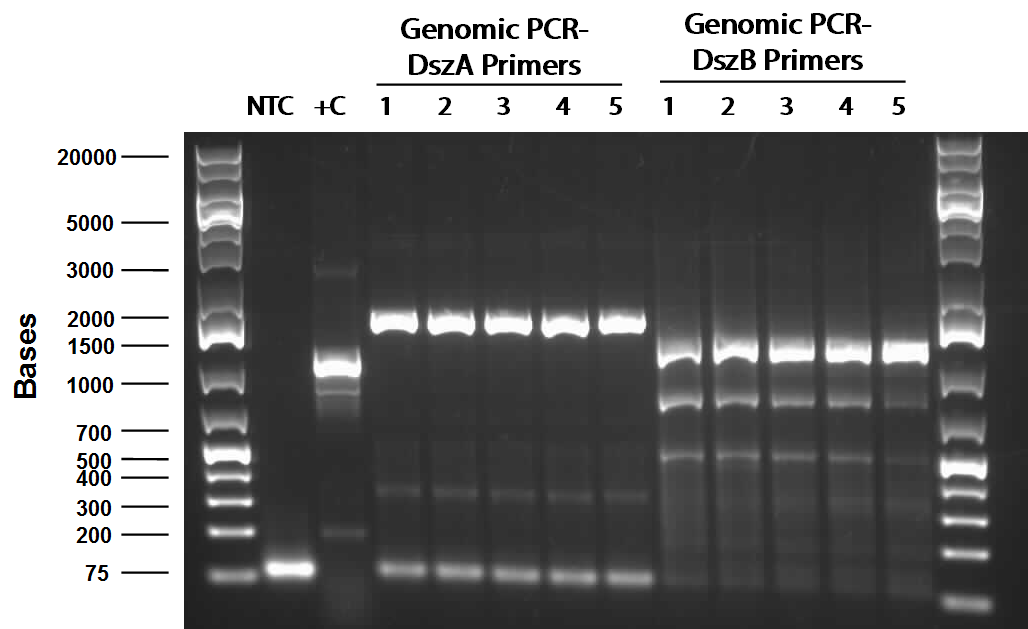
| - | Following the above successes with hpaC, the arrival of our | + | Following the above successes with<i> hpaC</i>, the arrival of our <i>Rhodococcus</i> strain afforded us the opportunity to begin investigation of the Dsz operon using the primers current in our possession. This strain is an environmental isolate that has been shown by <b> someone</b> to be an active desulfurizer. The gram-positive nature of the strain also dictated we explore various lysing strategies before the genes encoding the Dsz enzymes could be amplified for further purification and biobrick construction steps. PCR was carried out using <i>dszA</i> primers on three different treatments {microwave, lysate buffer, and a control} which yielded banding pattern around 1200 base pairs for the lysate treatment (2%SDS and 10% tritonX-100, plus heat for 5mins at 98C). |
| + | <b> DO WE HAVE A PICTURE</b></p> | ||
<h2>Week 6 (June 4 - June 8)</h2> | <h2>Week 6 (June 4 - June 8)</h2> | ||
| Line 141: | Line 154: | ||
<p> We started off this week by determining the DNA concentration of our genomic prep samples from last week using the nanodrop. DNA concentration for all three putida strains was at least 1000 ng/microlitre, well above what was needed for PCR. 1/2 and 1/3 dilutions were prepared for all three strains so as not to have an excess of template DNA in PCR reactions. PCR was performed on all 3 strains using primers for CarAc, CarAd, AntA, AntB, and AntC using 6 replicates per gene. The only successful amplification appeared to be AntB and CarAc, both from strain 28 (with weaker bands in strain 29). We then performed another PCR, just on those two genes with an increased amount of Taq polymerase to hopefully get enough amplified DNA to move forward with. This resulted in strong bands for both at the expected size. We then performed PCR purification using the Qiaquick kit and obtained samples containing 33.5 ng/microlitre of AntB DNA and 129 ng/microlitre of CarAc DNA. These concentrations were both sufficient to begin a restriction digest and ligation of these parts into vector PSB1C3. Next week we hope to verify the results of the restriction digest, continue to amplify CarAc and AntB from strain 28, and hopefully submit a biobrick for sequencing.</p> | <p> We started off this week by determining the DNA concentration of our genomic prep samples from last week using the nanodrop. DNA concentration for all three putida strains was at least 1000 ng/microlitre, well above what was needed for PCR. 1/2 and 1/3 dilutions were prepared for all three strains so as not to have an excess of template DNA in PCR reactions. PCR was performed on all 3 strains using primers for CarAc, CarAd, AntA, AntB, and AntC using 6 replicates per gene. The only successful amplification appeared to be AntB and CarAc, both from strain 28 (with weaker bands in strain 29). We then performed another PCR, just on those two genes with an increased amount of Taq polymerase to hopefully get enough amplified DNA to move forward with. This resulted in strong bands for both at the expected size. We then performed PCR purification using the Qiaquick kit and obtained samples containing 33.5 ng/microlitre of AntB DNA and 129 ng/microlitre of CarAc DNA. These concentrations were both sufficient to begin a restriction digest and ligation of these parts into vector PSB1C3. Next week we hope to verify the results of the restriction digest, continue to amplify CarAc and AntB from strain 28, and hopefully submit a biobrick for sequencing.</p> | ||
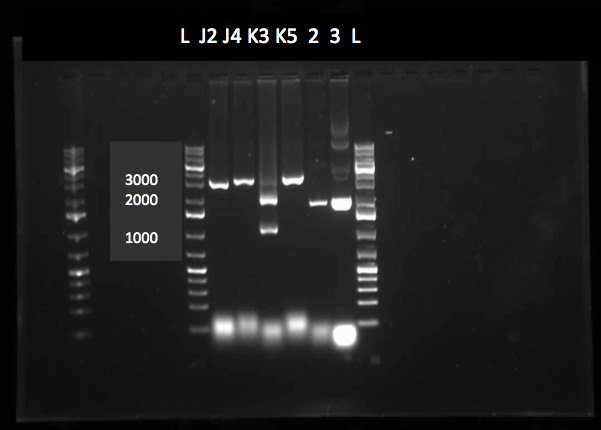
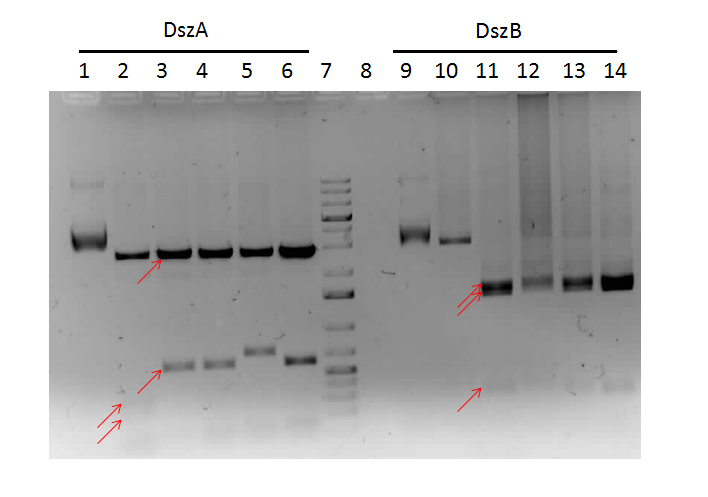
| - | <h3> | + | <h3>Desulfurization</h3> |
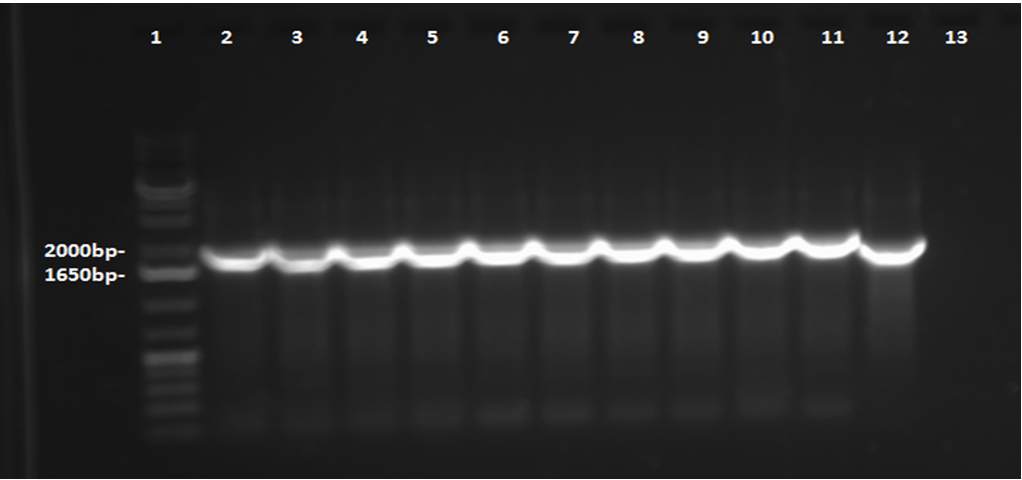
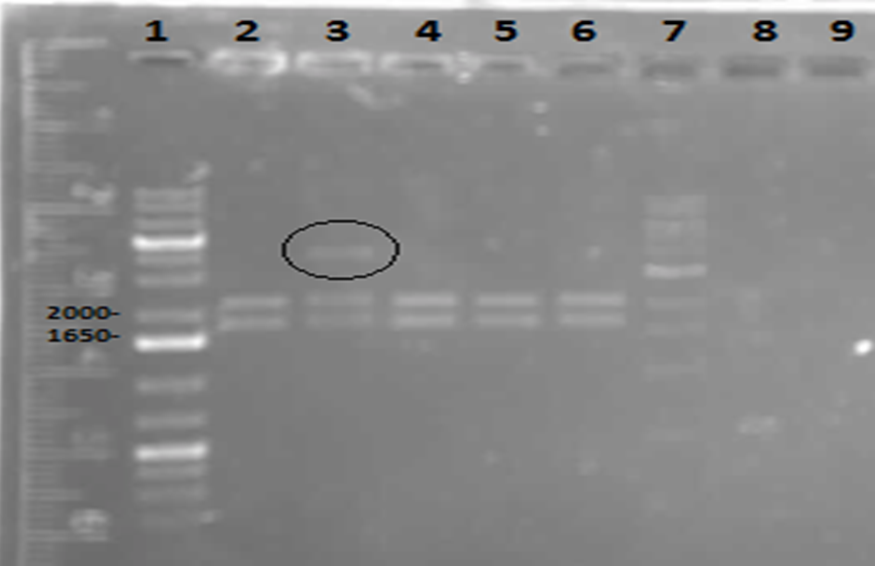
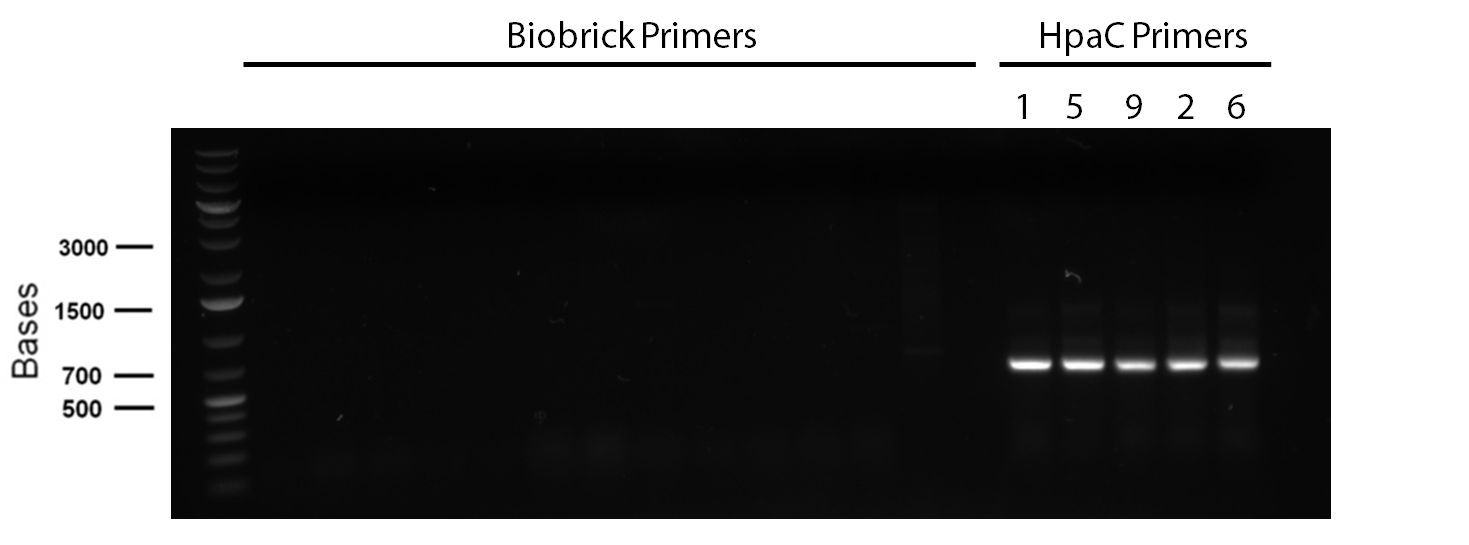
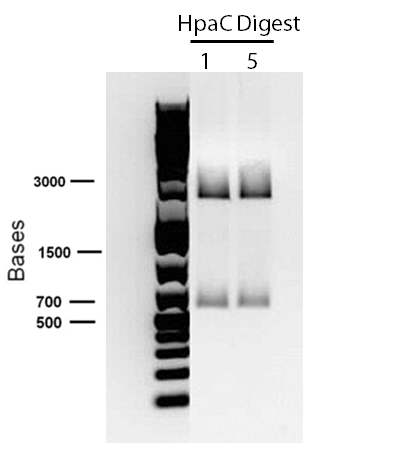
| - | < | + | <p> In order to confirm the <i>hpaC</i> biobrick construction, two sets of colony PCR were performed, choosing white colonies from the 3 plates we grew last week (white colonies indicate a loss of the RFP generator in the pSB1C3 backbone, and therefore allow for weeding out of the colonies which are simply the original plasmid vector). These reactions were carried out both with <i>hpaC</i> primers and with stanndard biobrick primers designed against the plasmid backbone. After running them on the gel we saw equal bands for the PCR reactions performed using <i>hpaC</i> primers <b>FIRST PIC</b>(However, a PCR using biobrick primers was performed later and the same results were obtained). Colonies 1(-) and 5(-) were used to make overnight cultures, which were then miniprepped the following day to obtain the plasmid DNA of the putative <i>hpaC</i> biobrick. Digestions were performed on the miniprep products using EcoRI and Pst to look for part size as further verification for the genes presence in the plasmid. The results were good and two bands were observed on each column (one for vector and the other for hpaC)(<b>second figure</b>). <i>hpaC</i> was sent in for sequencing. </p> |
| + | <p> | ||
| - | + | [[File:UCalgary2012_04.06.2012-desulfurisation_hpacverification.jpg|thumb|500px|center]] | |
| - | [[File:Ucalgary2012 06.06.2012-digestion of hpaC with E and P.jpg|thumb|500px|center]]<html> | + | [[File:Ucalgary2012 06.06.2012-digestion of hpaC with E and P.jpg|thumb|500px|center]]<html></p> |
| - | </ | + | |
| - | <p> PCR reagents were prepared to re-test/confirm previous results of dszA amplification following two different lysing treatments (microwave + lysate buffer). This time, all three genes were amplified and gel verification showed clear banding patterns around 500bp range for all three genes for the microwave treatment. Remaining PCR products were run on a gel and extracted for further purification steps; however, presence of any genetic material were not confirmed through nanodropping which raised concerns about the composition of the purified products | + | |
| + | <p> PCR reagents were prepared to re-test/confirm previous results of <i>dszA</i> amplification following two different lysing treatments (microwave + lysate buffer). This time, all three genes were amplified and gel verification showed clear banding patterns around 500bp range for all three genes for the microwave treatment.<b> WHY DID WE PROCEED WITH THIS THAT ISNT THE RIGHT SIZE</b> Remaining PCR products were run on a gel and extracted for further purification steps; however, presence of any genetic material were not confirmed through nanodropping which raised concerns about the composition of the purified products, the success of the initial amplification step, or perhaps even the lysis treatment. Further experimentation will have to be carried out to troubleshoot.</p> | ||
<h2>Week 7 (June 11 - June 15)</h2> | <h2>Week 7 (June 11 - June 15)</h2> | ||
| Line 173: | Line 187: | ||
<h3>Denitrification</h3> | <h3>Denitrification</h3> | ||
<p>The purified PCR products for CarAc and AntB were digested with restriction enzymes EcoRI+SpeI, EcoRI+PstI, and XbaI+PstI. However, only PSB1C3 plasmids that had been digested with EcoRI+SpeI and EcoRI+PstI were available to attempt ligation. Gel results were inconclusive on the restriction digest product as the plasmid size appeared much larger than expected. However, transformation was still attempted on the ligation products for both genes (both using EcoRI+SpeI ligation into PSB1C3 plasmid). The transformation products were plated onto chloro plates to select for colonies that had the chloro resistance gene on the PSB1C3 plasmid. Also this week another round of PCR was performed for the Car and Ant genes, however all, but CarAc showed bands in the negative control lane, possibly indicating contamination or the formation of primer dimers. The CarAc bands were fairly weak and a PCR purification resulted in very low DNA concentration, insufficient to move onto restriction digest. Next week's plan hinges heavily on the result of the transformation from this Friday. </p> | <p>The purified PCR products for CarAc and AntB were digested with restriction enzymes EcoRI+SpeI, EcoRI+PstI, and XbaI+PstI. However, only PSB1C3 plasmids that had been digested with EcoRI+SpeI and EcoRI+PstI were available to attempt ligation. Gel results were inconclusive on the restriction digest product as the plasmid size appeared much larger than expected. However, transformation was still attempted on the ligation products for both genes (both using EcoRI+SpeI ligation into PSB1C3 plasmid). The transformation products were plated onto chloro plates to select for colonies that had the chloro resistance gene on the PSB1C3 plasmid. Also this week another round of PCR was performed for the Car and Ant genes, however all, but CarAc showed bands in the negative control lane, possibly indicating contamination or the formation of primer dimers. The CarAc bands were fairly weak and a PCR purification resulted in very low DNA concentration, insufficient to move onto restriction digest. Next week's plan hinges heavily on the result of the transformation from this Friday. </p> | ||
| + | |||
<h3>Desulfurisation</h3> | <h3>Desulfurisation</h3> | ||
| - | <p> This week, we | + | <p> This week, we focused on amplifying <i>dsz</i> genes from our <i>Rhodococcus</i> strain for construction into biobricks. We also wanted to purify the PSB1C3-<i>hpaC</i> and pUC18-<i>hpaC</i> plasmids to replenish our current stocks. For the <i>dsz</i> aspect, we were able to successfully grow extra plates of <i>Rhodococcus</i> strain which was used to inoculate PCR tubes. The PCR did not go well, with significant streaking and false positives with similar banding pattern to previous gels run in the previous week. A final gel verification of a random sample of a tube of PCR products from dszA,B,C respectively and two negative control treatments involving master mix only and the lysed cells only illustrated the lack of discrepancy between the supposed successful amplification and the lysed cells (with lysate buffer) alone. Because of this we decided to take a different approach involving plasmid isolation carried out before PCR, rather than applying the PCR reagents directly to a lysed culture sample.</p><p> PSB1C3-hpaC verification through sequencing was successful, confirming the construction of our first biobrick. Subsequently, O/N cultures of the plasmid containing cultures were prepared and stored in glycerol at -80C. Furthermore, verification of catalase gene part (KatG-LAA, BBa_K137068)sent as a culture stab from the parts registry was initiated, with our newly identified biobricked-hpaC acting as a positive control, but the banding pattern was not very conclusive. </p> |
<h2>Week 8 (June 18 - June 22)</h2> | <h2>Week 8 (June 18 - June 22)</h2> | ||
| Line 192: | Line 207: | ||
<h3>Desulfurisation</h3> | <h3>Desulfurisation</h3> | ||
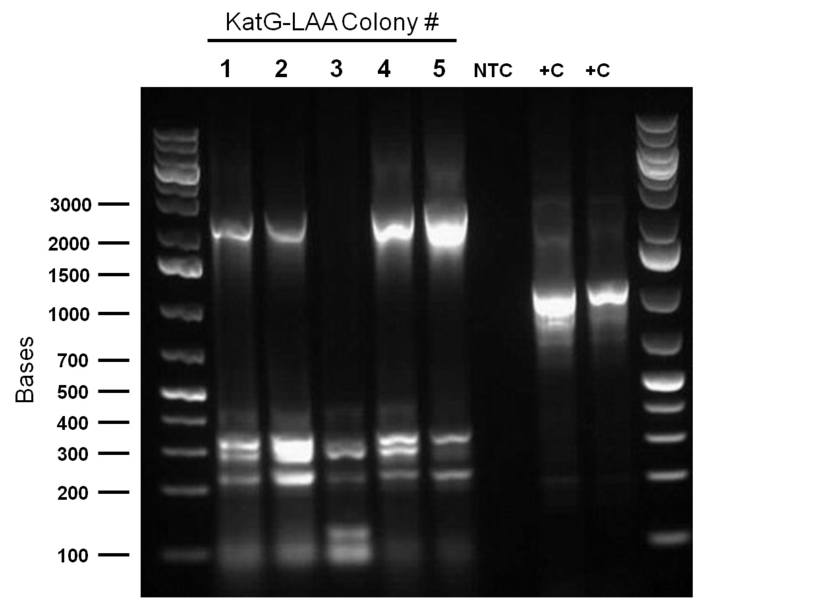
| - | <p>PCR was reattempted on | + | <p>PCR was reattempted on <i>Rhodoccocus</i> that was lysed using two different dilutions of the lysate buffer, but the gel verification confirmed the previous failure in using this approach. An alternative that involved preparation of an overnight culture of the <i>Rhodococcus</i> cells followed by a plasmid purification was followed. The plasmid purification eventually yielded plasmid samples with concentrations of 98.6ng/uL to 182.7ng/uL (4 samples obtained overall). Additionally, the catalase biobrick was used to transform some stock competent cells, and samples of some colonies were subsequently PCR'ed. Although, the gel verification showed some potential contamination, and the required banding patterns at around 2200bp was not obtained. </p> |
<h2>Week 9 (June 25 - June 29)</h2> | <h2>Week 9 (June 25 - June 29)</h2> | ||
| Line 203: | Line 218: | ||
<h3>Desulfurization</h3> | <h3>Desulfurization</h3> | ||
| - | <p> PCR was attempted to amplify the dsz operon utilising an adapted PCR protocol with | + | <p> PCR was attempted to amplify the genes of the <I>dsz</i> operon utilising an adapted PCR protocol with purified <i>Taq</i> polymerase that had been isolated from the host organism. Eventually, some banding pattern was obtained between 1200 and 1500 base pairs when a gradient thermocycler was used with melting temperatures ranging betweeen 55C to 65C. This was assumed to be indicative of successful amplification of <i>dszB</i>; however, further purification and gel verification results were inconclusive and no yield was obtained when placed tested using a nanodrop machine. |
<h3>Ring Cleavage</h3> | <h3>Ring Cleavage</h3> | ||
| Line 229: | Line 244: | ||
<h3>Desulfurization</h3> | <h3>Desulfurization</h3> | ||
| - | Top 10 E.coli cells were transformed | + | <p>Top 10 E.coli cells were transformed with R0011 (IPTG inducible promoter in psb1C3 backbone), and resulting colonies were tested using cPCR. |
| - | Colony PCR was performed on cells containing the catalase biobrick. Catalase is 2217bp long but since biobrick primers add about 200bp | + | Colony PCR was performed on cells containing the catalase biobrick. Catalase is 2217bp long but since biobrick primers add about 200bp, bands of 2400 bp were expected if the part was present in the biobrick. These bands were observed, indicating that the <i>KatG-LAA</i> gene was most likely present.</p> |
| - | dszA, dszB and dszC | + | <p></html>[[File:Ucalgary2012 4.7.2012 catalase colony pcr 2.jpg|thumb|500px|center]]<html></p> <p> |
| - | dszABC genes | + | |
| + | PCR using Phusion high fidelity polymerase was carried out on <i>dszA</i>, <i>dszB</i>, and <i>dszC</i> in a gradient thermocycler. Amplification of non-specific bands was present for <i>dszA</i> and <i>dszB</i>, however strong banding for the desired size of the gene was observed for both (around 1500 for <i>dszA</i>, 1100 for <i>dszB</i> </p> | ||
| + | |||
| + | <p></html>[[File:Ucalgary2012 6.7.2012.dszABphusionPCR.jpg|thumb|500px|center]]<html> | ||
| + | </p> <p> | ||
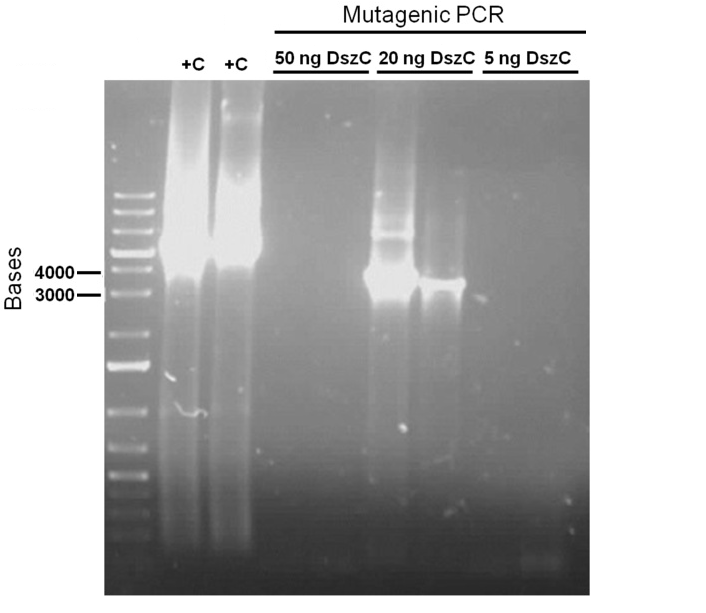
| + | Examining the sequences of the <i>dszABC</i> genes led to the discovery that all 4 had multiple illegal enzyme cut-sites in them that we have to eliminate before biobrick composite part construction can occur. <i>dszA</i> has four PstI cut sites, <i>dszB</i> has a PstI and a NotI and <i>dszC</i> has a PstI cut site. In order to eliminate cut sites, the Stratagene QuikChange mutagenesis procedure is going to be used, with the only alteration being that Kapa HiFi polymerase would be used during the process. Primers needed for the mutagenesis were designed based on the procedure mentioned above. </p> | ||
| - | |||
| - | |||
| - | |||
<h3>Ring Cleavage</h3> | <h3>Ring Cleavage</h3> | ||
| Line 258: | Line 275: | ||
<h3>Desulfurization</h3> | <h3>Desulfurization</h3> | ||
| - | <p> Following successful amplification of the dsz operon in the previous | + | <p> Following successful amplification of the <i>dsz</i> operon genes in the previous week, the genes were constructed into the PSB1C3 vector. Colony PCR verifications were observed to be positive. Furthermore, the insertion of part J13002 (pTetR)in front of the previously biobricked <i>hpaC</i> was attempted. Overnight cultures were also prepared using two colonies each for J13002 and R0011 (an IPTG inducible promoter that we hope to build in front of B0034). These cultures were then miniprepped to yield the respective parts.</p> |
| + | <p><b> PICTURE </b></p> | ||
| - | <p>Additionally, katG was built into a PSB1C3 backbone. The construction and availability of all these parts will be critical in the construction of our overall circuit for biodesulfurization. | + | <p>Additionally, <i>katG-LAA</i> was built into a PSB1C3 backbone. The construction and availability of all these parts will be critical in the construction of our overall circuit for biodesulfurization. Colonies which looked good on cPCR were used to prepare overnight cultures, and were miniprepped and sent in for sequencing verification the following day. |
| - | On the side, M9 minimal media was also prepared to carry out growth experimentation and overall desulfurization capability of | + | On the side, M9 minimal media was also prepared to carry out growth experimentation and overall desulfurization capability of </i>Rhodococcus</i> when exposed to DBT. The various growth treatments were M9 Media and glucose only, M9+glucose+DBT, M9+glucose+MgSO4+/-DBT, M9+glucose+MgCl2+/-DBT. 0.008g of FeCl2.4H2O was also added to each of the tubes. Samples were then inoculated with colonies of the <i>Rhodococcus</i>.</p> |
| Line 298: | Line 316: | ||
<h3>Desulfurization</h3> | <h3>Desulfurization</h3> | ||
| - | <p>This week, while awaiting sequencing verification results | + | <p>This week, while awaiting sequencing verification results which were required before we could begin the construction process, the desulfurization team initially aided in some of the tasks related to the other hydrocarbon groups. The success of the construction of BBa_J13002 with hpaC was also explored by using forward and reverse primers of R0040 (the promoter component of the composite part J13002). However, the eventual gel verification was inconclusive and sequencing results finally indicated an unsuccessful ligation. Additionally, the minimal media M9 preparation had been contaminated in the previous effort so this process was repeated to create tubes of each of the growth condition treatments detailed previously, and two repeats, one with an extra filtration step and one without was used to prepare the cultures.</p> |
<h2>Week 13 (July 23 - July 27)</h2> | <h2>Week 13 (July 23 - July 27)</h2> | ||
| Line 308: | Line 326: | ||
<h3>Desulfurization</h3> | <h3>Desulfurization</h3> | ||
| - | <p> Mutagenic primers were redesigned after initial design was found to have premature stop codons. | + | <p> Mutagenic primers were redesigned after initial design was found to have premature stop codons. As part of the redesign process in constructing our overall gene circuits for desulfurization, a backbone switch of R0011 into a chlor-resistant vector was necessary, so this was attempted. The subsequent transformed products were plated on a Chlor plate and selected colonies were used to prepare O/N cultures, then miniprepped before finally being digested with enzymes EcoRI and PstIA. The resulting gel verification images were inconclusive as they did not show the required banding pattern around 50bp. Meanwhile, colony PCR was run on colonies transformed with <i>katG-LAA</i> constructed into a PSB1C3 backbone, as well as the <i>hpaC</i>+J13002 construct. <i>katG</i> was shown to have been successfully amplified, verifying its presence in the colonies, so overnight cultures were prepared and subsequently miniprepped. On the other hand, the construct was not successful so a third attempt was carried out. Colony PCR treatments that used either R0011 forward primers or B0034 primers were used and the overall constructs were made either on a chlor-resistant, or amp-resistant vector. Preliminary images of the gel verification appeared to have confirmed the construct, although sequencing verification will be the final indicator of overall success. </p> |
<h3>Denitrification</h3> | <h3>Denitrification</h3> | ||
| Line 316: | Line 334: | ||
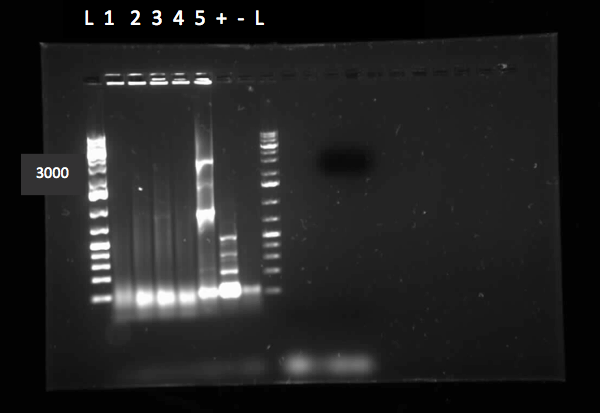
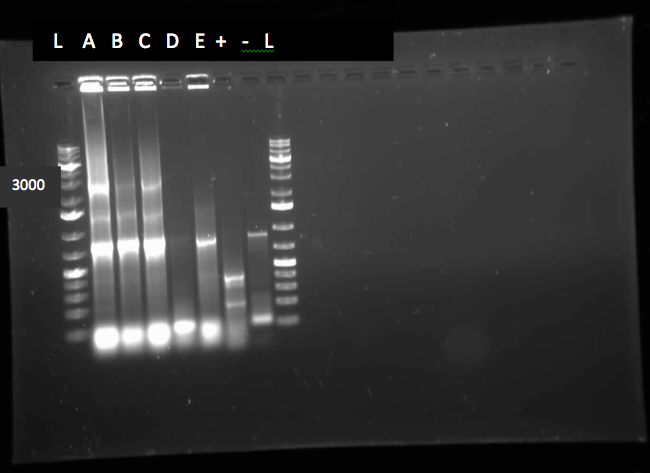
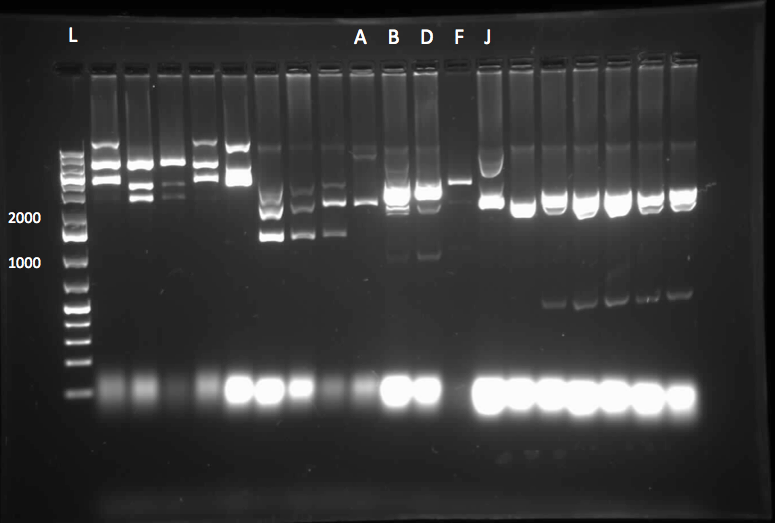
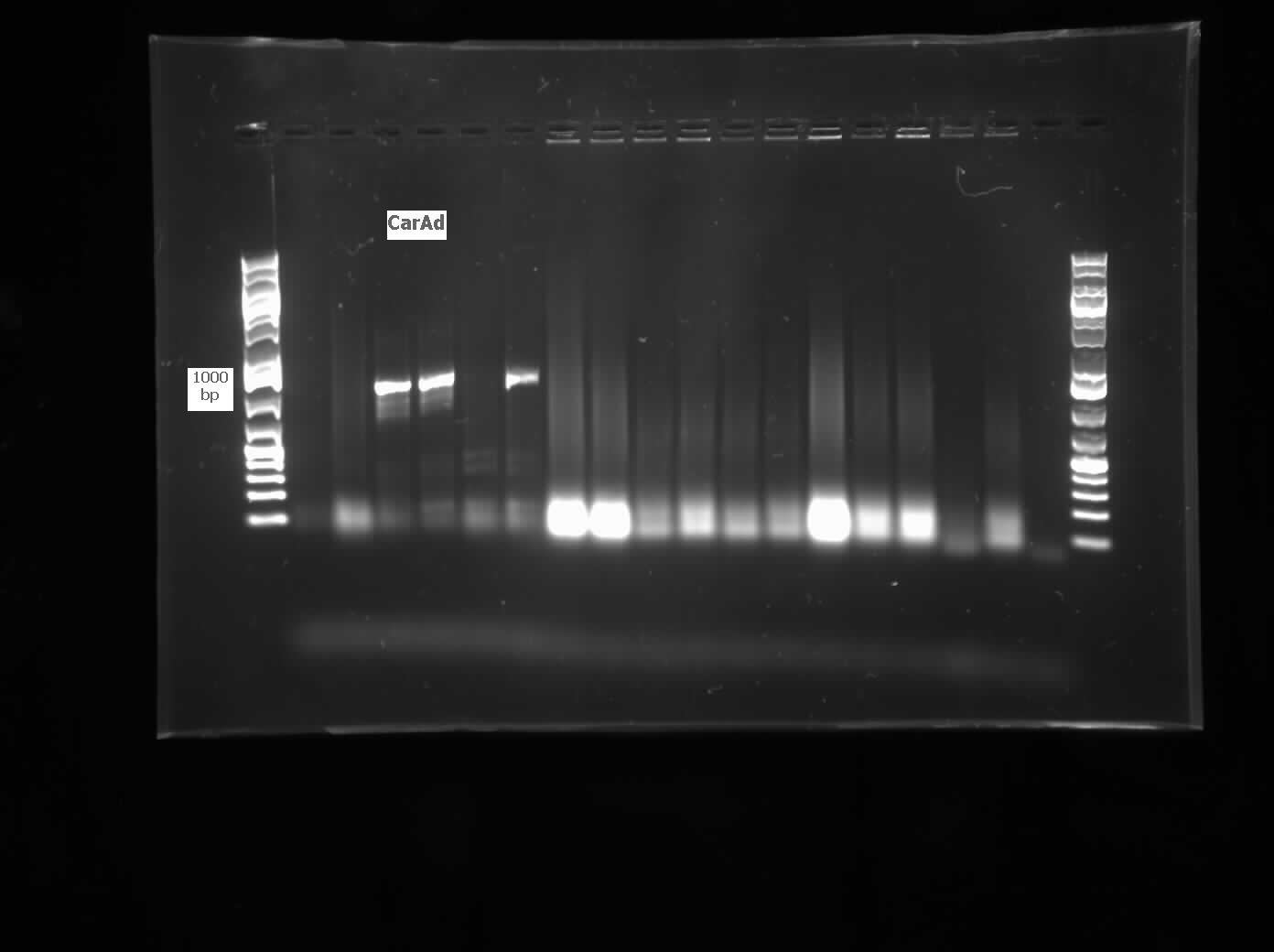
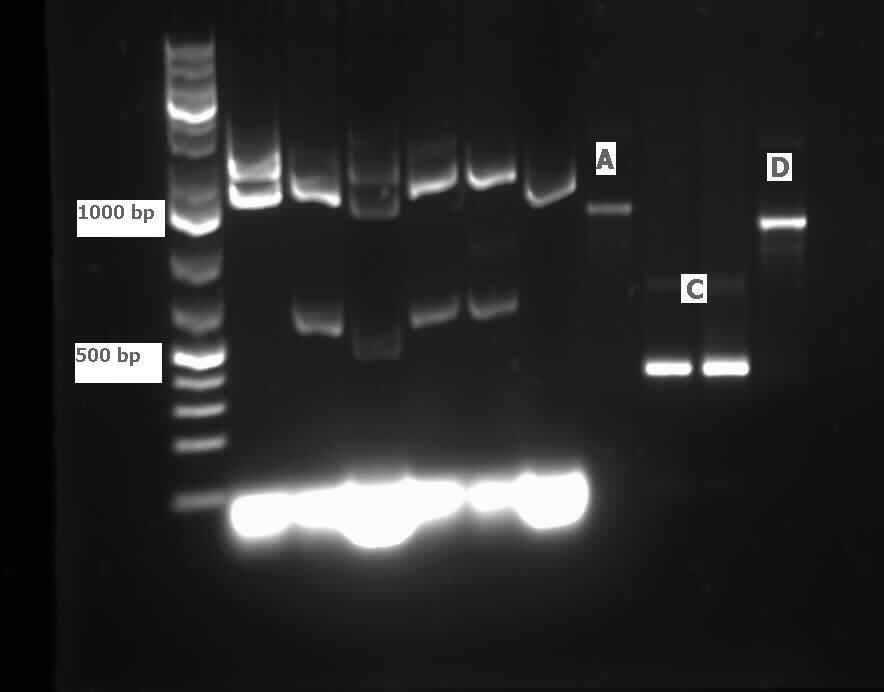
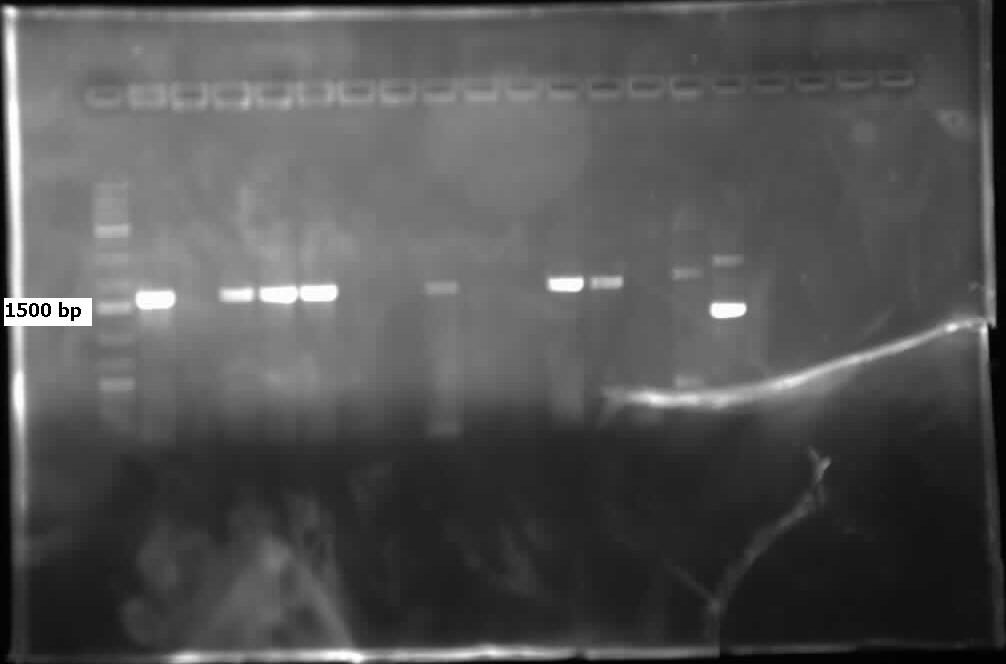
</html>[[File:07.23.2012.Restriction Digest of Car genes.jpg|500px|thumb|Gel of restriction digest of Car genes. A refers to CarAa, C to CarAc, and D to CarAd. Ladder shows that after purification and digestion DNA is still the correct size.|center]] | </html>[[File:07.23.2012.Restriction Digest of Car genes.jpg|500px|thumb|Gel of restriction digest of Car genes. A refers to CarAa, C to CarAc, and D to CarAd. Ladder shows that after purification and digestion DNA is still the correct size.|center]] | ||
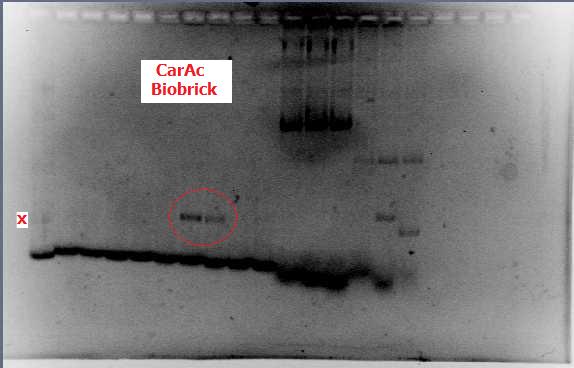
[[File:CarAc biobrick confirmation PCR.jpg|500px|thumb|Gel of confirmation PCR done on competent Top 10 E. coli cells in which a PSB1C3 vector containing the CarAc gene had been inserted. Red X marks the 500 bp mark on the weak ladder showing the product to be the expected size (323 bp gene + 2*100 bp biobrick sites on each end).|center]] <html> | [[File:CarAc biobrick confirmation PCR.jpg|500px|thumb|Gel of confirmation PCR done on competent Top 10 E. coli cells in which a PSB1C3 vector containing the CarAc gene had been inserted. Red X marks the 500 bp mark on the weak ladder showing the product to be the expected size (323 bp gene + 2*100 bp biobrick sites on each end).|center]] <html> | ||
| + | |||
| + | <h3>Catechol Degradation</h3> | ||
| + | |||
| + | <p>The colony PCR of part J33204 (XylE with a native rbs site) was done again and all five colonies looked like they contained the plasmid after running the products on a gel. After miniprepping and running a restriction digest on these five colonies two of them were sent to sequencing. | ||
| + | An assay using the compound catechol was also started this week. E.coli cells (top 10) transformed with the part K118021 (XylE with a Pcst promoter) were used in this assay. Six overnight cultures were made, 3 using M9-MM and 3 using LB. These were spun down and resuspended in fresh M9-MM and LB and brought to a concentration of 0.1 M catechol by using a 1M stock solution. There seemed to be an initial colour change in the reactions from clear to yellow which was only visible in the cultures using M9-MM. All of the cultures were incubated overnight and they turned a black green colour. For this assay we should have used the supernatant because the reaction takes place outside of the cell so this is what we did next week.</p> | ||
<h2>Week 14 (July 30 - August 3)</h2> | <h2>Week 14 (July 30 - August 3)</h2> | ||
| + | |||
| + | |||
| + | <h3>Desulfurization</h3> | ||
| + | <p>Sequencing results from the previous week's constructs were available confirming the contruction of KatGLAA in a chlor-resistant backbone, however, the backbone switch of R0011 also to PSB1C3 was not successful, probably owing to the small length of the RBS. On the other hand, the construction of J13002+hpaC was finally sent in for sequencing too, pending results. Site-directed mutagenesis of the dsz operon was also initiated - dszA has four PstI cut sites. dszB has a PstI and a NotI site. dszC has two PstI cut sites. Mutagenesis was started this week to change a single base pair in these genes in a way that the cut site is eliminated but the amino acid the gene codes for stays intact. In addition another mutagenesis has to be performed in order to replace the Tyr at position 63 of dszB gene with a Phe. Ohshiro 2007 showed that this mutation increases the activity of the enzyme. | ||
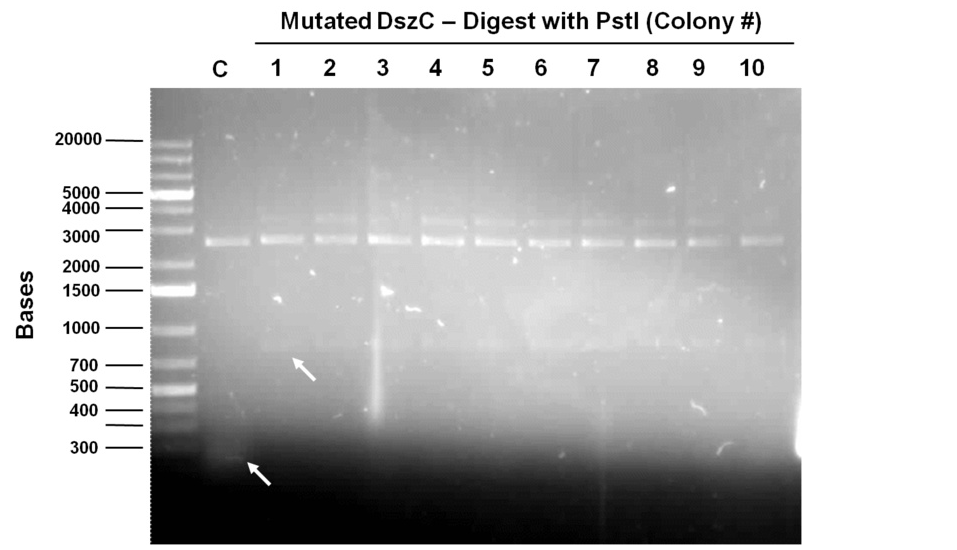
| + | The mutagenesis was started by mutating the second PstI site in dszC (PstI2) and a control plasmid containing a point mutated β-galactosidase gene was also mutated with its specific primers. Kappa Hifi kit was used for all the mutagenesis. After the PCR mutagenesis and running a part of the PCR products on the gel, amplification was observed. Then the PCR products were DpnI digested to degrade the parental DNA and they were transformed. Control PCR products were plated on an amp plate containing IPTG and X-gal. The colonies that grew on the control plates were blue indicating that the mutagenesis had worked and the β-galactosidase gene was now functional. Then O/N culture of dszC mutants were miniprepped and digested with PstI enzyme and the results were successful<b> (Lisa gel pic)</b>.</p><p> | ||
| + | Attempts to do all the mutations in the genes in one step using the Knight procedure failed (http://openwetware.org/wiki/Knight:Site-directed_mutagenesis/Multi_site). This procedure used Taq ligase buffer. We suspected that the reason this procedure is not working might be that the Kappa polymerase is not functional in Taq ligase buffer. Therefore we did some experiments on the controls in Taq ligase kit and kappa polymerase kit to find out the buffer that they both work best in. The result was that both enzymes (kappa polymerase and Taq ligase) work best in a buffer made of half Taq ligase buffer and half kappa polymerase buffer. </p> | ||
| + | |||
| + | </html>[[File:Ucalgary2012 31.7.2012 dszc mutagenesis 5 20 and 50ng.jpg|thumb|500px|center]] | ||
| + | [[File:UCalgary 02.08.12 dszC psti digest mutagenesis.jpg|thumb|500px|center]]<html> | ||
| + | </html> | ||
<h3>Decarboxylation</h3> | <h3>Decarboxylation</h3> | ||
| Line 335: | Line 368: | ||
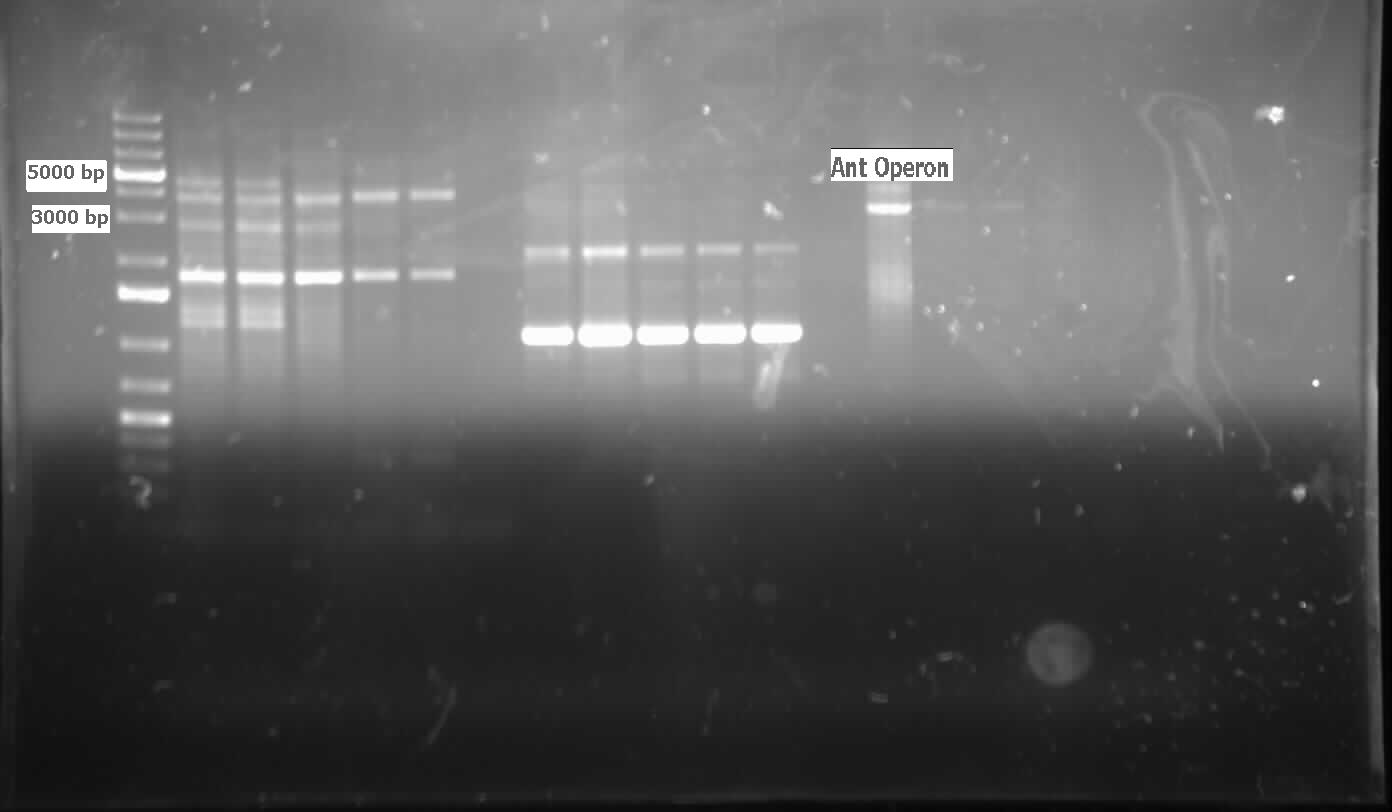
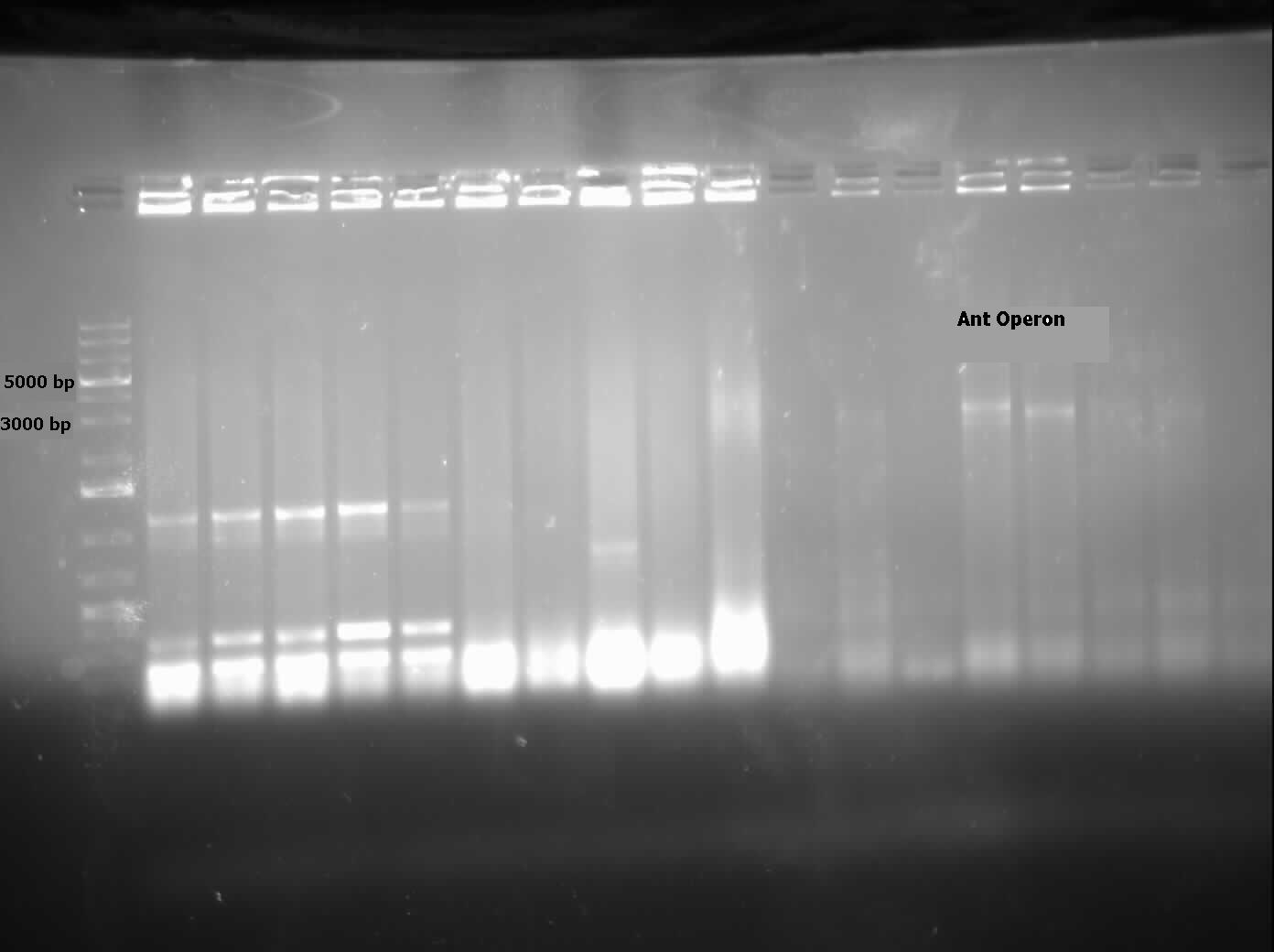
[[File:07.31.2012.Ant operon.jpg|500px|thumb|Ant operon amplified from genomic prep of Pseudomonas LD2 at the expected size of 3000 bp.|center]] | [[File:07.31.2012.Ant operon.jpg|500px|thumb|Ant operon amplified from genomic prep of Pseudomonas LD2 at the expected size of 3000 bp.|center]] | ||
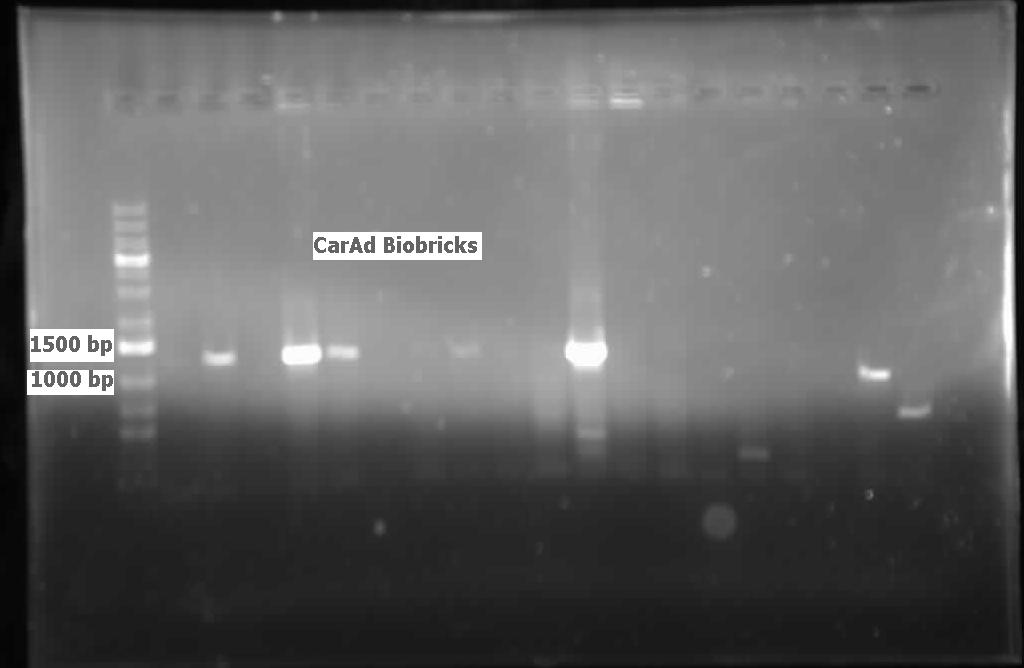
[[File:08.01.2012 CarAdBB.jpg|500px|thumb|Lanes 2,4,5, and 11 contain CarAd genes amplified from colony PCR (using biobrick primers) of the PCR product that had been ligated into PSB1C3 and transformed into competent E.Coli.|center]] | [[File:08.01.2012 CarAdBB.jpg|500px|thumb|Lanes 2,4,5, and 11 contain CarAd genes amplified from colony PCR (using biobrick primers) of the PCR product that had been ligated into PSB1C3 and transformed into competent E.Coli.|center]] | ||
| - | [[File:08.01.2012 CarAaBB.jpg|500px|thumb|Lane 9 contains CarAa genes amplified from colony PCR (using biobrick primers) of the PCR product that had been ligated into PSB1C3 and transformed into competent E.Coli. Although larger than expected, the positive control RFP generator in lane 10 is also larger than its expected size (~1200 bp including biobrick bookends) indicating that the gel may have a slant in it.|center]] | + | [[File:08.01.2012 CarAaBB.jpg|500px|thumb|Lane 9 contains CarAa genes amplified from colony PCR (using biobrick primers) of the PCR product that had been ligated into PSB1C3 and transformed into competent E.Coli. Although larger than expected, the positive control RFP generator in lane 10 is also larger than its expected size (~1200 bp including biobrick bookends) indicating that the gel may have a slant in it.|center]]<html> |
| + | |||
| + | <h3>Catechol Degradation</h3> | ||
| + | |||
| + | <p>The sequencing results for the part J33204 matched the part so a construction was started. The construction was a 3-way ligation putting the tetR promoter (R0040) and XylE (J33204) into a pSB1C3 vector. The ligation product was transformed into competent cells and a colony PCR was done on the transformants using both bio brick and R0040 primers. | ||
| + | The primers for XylT, which is a ferredoxin that allows for XylE to be activated after a reaction with catechol, arrived and a colony PCR on four different strains of Pseudomonas putida was done. None of the strains seemed to contain the TOL plasmid, which is where XylT is found in certain strains of P. putida, because no bands showed up in the gel. | ||
| + | The catechol assay was continued this week. Six overnight cultures were made, 3 using M9-MM and 3 using LB. They were spun down and the supernatant was brought to a catechol concentration of 0.1M, 0.2 M and 0.5 M using a 1M stock solution. The solutions turned light yellow initially, but after a few hours they turned brown. | ||
| + | The assay was repeated with the same procedure as above but a control, a colony that did not contain the part K118021, was added. After the catechol was added to the supernatants there was a colour change from clear to yellow in all of the tubes, even the control. For a negative control catechol was added to M9-MM and this solution became pink. The tubes were incubated on the bench and checked regularly. The colour changed from yellow to pink to brown when they were left on the bench overnight.</p> | ||
<h2>Week 15 (August 7 - August 10)</h2> | <h2>Week 15 (August 7 - August 10)</h2> | ||
| + | |||
| + | <h3>Desulfurization</h3> | ||
| + | <p>Sequencing results for J13002/hpaC returned negative, so a 3-way ligation method was used to retry the construction. The following parts were ligated with the restriction enzymes indicated in brackets after each: J13002(E/S) + HpaC (X/P) + PSB1K3 (E/P). Also, the more conventional construction of J13002(S/P) + HpaC(X/P) was reattempted. Furthermore, 3-way ligations were also attempted for B0034+KatGLAA+PSB1K3, and R0011+B0034+PSB1C3, as well as the two-way contruction of just KatGLAA after the B0034. After plating these transformations, colony PCRs were carried out on selected colonies and various samples that gave an indication on the gels of being successful were used to prepare O/N cultures then subsequently miniprepped.<b>(insert gel images!)</b> With regards to the site-directed mutagenesis side of the experimentation, dszA-PstI1 (the first PstI cut site in dszA) , dszB-PstI and dszC(PstI2 mutated)-PstI1 mutagenesis were performed following the procedure explained in the previous week. The gel below shows the successful result of digest confirmation. | ||
| + | Multisite mutagenesis was repeated again using the modified buffer (half Taq ligase buffer and half Kappa buffer). However it was not successful again. We also tried doing multisite mutagenesis using pfu Turbo polymerase and following the Knight procedure without any modifications. No successful results were observed. </p> | ||
| + | |||
| + | </html> | ||
| + | [[File:Ucalgary2012 Digestion confirmation of mutagenesis in dszAPstI1, dszBPstI and dszCPstI1 (PstI2 site mutated)..jpg|thumb|500px|center]]<html> | ||
| + | </html> | ||
| + | |||
| + | <h3>Decarboxylation</h3> | ||
| + | |||
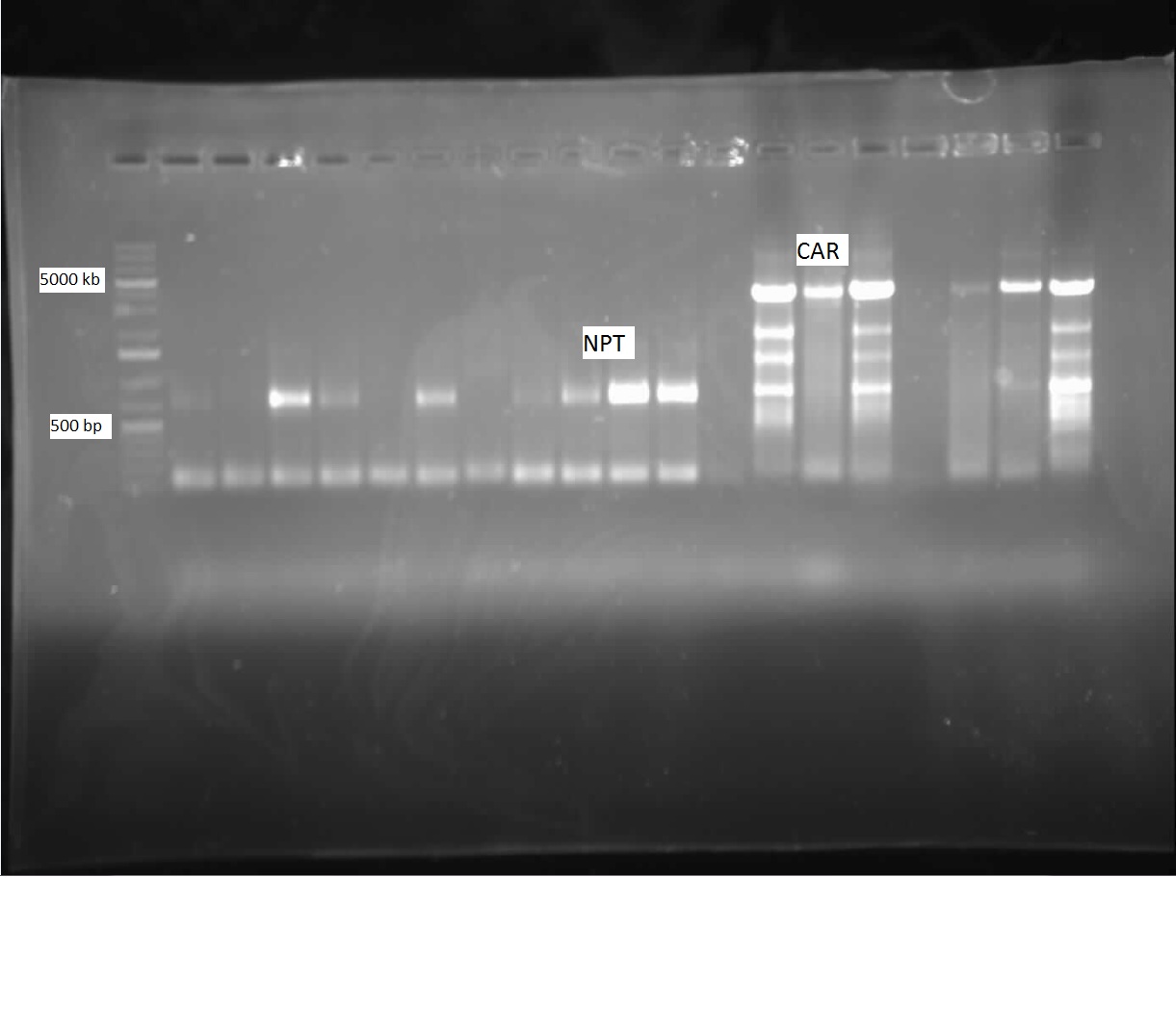
| + | <p> We ran the gel on the CAR and NPT colony PCRs from last week, but unfortunately these gels were blank. Another colony PCR was performed, with 12 colonies from each of the following plates: CAR/PET, NPT(X/P), NPT(E/P). Both NPT plates did not look very promising, as they were completely covered by red colonies, with only a few white ones. Many of the white colonies from the previous week had become red. 8 overnight cultures were also performed so that a restriction digest could be done on the CAR colonies, to see if the part was there. These overnight cultures were prepared with the Kan antibiotic. Another ligation was done with the three combinations of cut inserts and vectors: CAR with PET, NPT (E/P) with PSB1C3, and NPT (X/P) with PSB1C3. Restriction digests were performed on the CAR overnight colonies with AscI and HindIII, and when the gel was run, a single lane was shown to contain both the CAR gene and PET vector, as shown below:</p> | ||
| + | |||
| + | </html>[[File:CARinPET.jpg|thumb|500px|center]]<html> | ||
| + | |||
| + | <p>As shown in the gel above, lane 3 indicates 2 bands, the uppermost at about 5.4kb, identifying as PET. This single lane also contains a clear band of CAR, a bit under the 5000bp mark, at 4690bp. The ladder used was 1 kb plus. This successful result allowed us to proceed with mutagenesizing the 6 illegal cut sites of CAR, including: 2 PstI sites, 1 EcorI site, and 3 NotI sites, with the primers that we had designed. Two additional colony PCRs of NPT were performed, but unfortunately a band indicating NPT was not visible in any of the traisl, even when an additional plate of both the NPT (E/P) and NPT (X/P) restriction digests were ligated and transformed. Very few white colonies could be seen on any of the transformations, but we were determined to continue trying. We then attempted to mutagenesize the first illegal cut site of CAR, which would be a PstI site. The mutagenesis primers needed to be diluted and stock solutions were made. Then, we attempted to mutagenesize the PstI site of CAR with that site's specific primers, using various concentrations of template DNA (5ng, 20ng, 50ng). We used the KAPA system to perform this plasmid amplification, using the thermocycler settings of the Desulfurization team. The mutagenesis PCR would be run over the weekend to hopefully have successful results. </p> | ||
| + | |||
| + | |||
| + | |||
<h3>Denitrification (or are we calling it denitrogenation now??)</h3> | <h3>Denitrification (or are we calling it denitrogenation now??)</h3> | ||
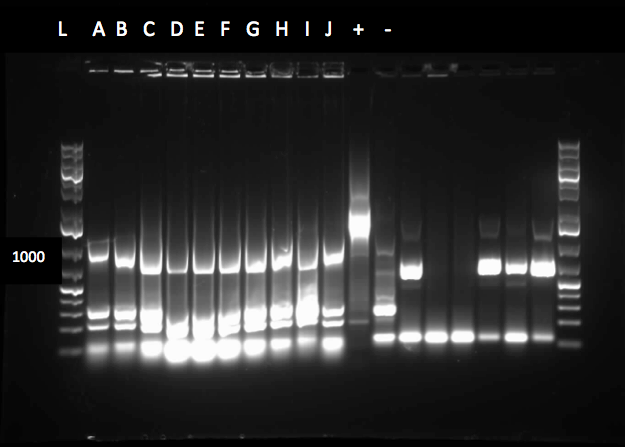
<p>This week we received the sequencing results for CarAa and CarAd. CarAd was a match, however CarAa did not match at all and had misplaced biobrick restriction enzyme sites. Due to this we decided to restart the biobricking process for CarAa all the way back to PCR from the plasmid. By the end of the week we had progressed to the point of transforming it into competent E. coli cells, however a thick lawn of bacteria had grown overnight making it impossible to pick colonies. A streak plate was made from this plate to attempt to get less dense growth. Since the sequencing came back positive for CarAd the next step was to perform site directed mutagenesis on its NotI restriction enzyme site using the primers that were designed previously. The PCR was successful, showing very bright bands at 3000 bp, and the PCR product was digested with DpnI to remove parental DNA. Transformation of this product will indicate whether the mutagenesis was successful. We were also able to successfully perform a genomic prep of Rhodococcus erythropolis and amplify the AmdA gene this week. It was ligated into PSB1C3 and transformed into competent E. coli by Friday, and a confirmation colony PCR was run on Friday evening to verify that the gene was inserted. These results will be available by Monday. After many attempts the Ant operon was finally ligated and transformed as well this week. Confirmation PCR showed bands of about 3000 bp which is consistent with the size of the operon. By Friday the successful colonies had been grown up and miniprepped, ready to send for sequencing early next week. The CarAc/B0034 construct was also successfully transformed this week and verified by running a PCR using the B0034 forward primer. It was also miniprepped and is ready to send for sequencing next week. In addition to this work constructing biobricks we also started an experiment to demonstrate the function of the pCAR plasmid in LD2. Strains of LD2 were grown up in LB and then washed and placed in B-N media + glucose and various experimental additions. These additions included carbazole, 4-PBAH (a nitrogen containing naphthenic acid), ammonia chloride, and a negative control with no nitrogen source. Strains of E. coli were also grown under the same conditions to compare growth and ammonia production of LD2 to a strain that does not contain the pCAR plasmid. These results should act as a preview of what we expect our final biobrick containing E. coli species to produce as well as allowing us to establish LD2 as a positive control for this experiment. </p> | <p>This week we received the sequencing results for CarAa and CarAd. CarAd was a match, however CarAa did not match at all and had misplaced biobrick restriction enzyme sites. Due to this we decided to restart the biobricking process for CarAa all the way back to PCR from the plasmid. By the end of the week we had progressed to the point of transforming it into competent E. coli cells, however a thick lawn of bacteria had grown overnight making it impossible to pick colonies. A streak plate was made from this plate to attempt to get less dense growth. Since the sequencing came back positive for CarAd the next step was to perform site directed mutagenesis on its NotI restriction enzyme site using the primers that were designed previously. The PCR was successful, showing very bright bands at 3000 bp, and the PCR product was digested with DpnI to remove parental DNA. Transformation of this product will indicate whether the mutagenesis was successful. We were also able to successfully perform a genomic prep of Rhodococcus erythropolis and amplify the AmdA gene this week. It was ligated into PSB1C3 and transformed into competent E. coli by Friday, and a confirmation colony PCR was run on Friday evening to verify that the gene was inserted. These results will be available by Monday. After many attempts the Ant operon was finally ligated and transformed as well this week. Confirmation PCR showed bands of about 3000 bp which is consistent with the size of the operon. By Friday the successful colonies had been grown up and miniprepped, ready to send for sequencing early next week. The CarAc/B0034 construct was also successfully transformed this week and verified by running a PCR using the B0034 forward primer. It was also miniprepped and is ready to send for sequencing next week. In addition to this work constructing biobricks we also started an experiment to demonstrate the function of the pCAR plasmid in LD2. Strains of LD2 were grown up in LB and then washed and placed in B-N media + glucose and various experimental additions. These additions included carbazole, 4-PBAH (a nitrogen containing naphthenic acid), ammonia chloride, and a negative control with no nitrogen source. Strains of E. coli were also grown under the same conditions to compare growth and ammonia production of LD2 to a strain that does not contain the pCAR plasmid. These results should act as a preview of what we expect our final biobrick containing E. coli species to produce as well as allowing us to establish LD2 as a positive control for this experiment. </p> | ||
| + | |||
| + | </html>[[File:08.07.2012 amdA pcr from genomic prep.jpg|500px|thumb|AmdA amplified from the genomic prep of R. Erythropolis shows bands just greater than 1500 base pairs in lanes 2,4-6,9, and 12-13. Lane 14 was a negative control. The expected size of AmdA is 1566 base pairs.|center]] | ||
| + | [[File:08.09.2012 confirmation pcr of ant operon.jpg|500px|thumb|The two labelled lanes show amplification at 3000 base pairs from a confirmation colony PCR using the biobrick primers. The expected size of the Ant operon with the biobrick bookends is about 3100 base pairs.|center]] | ||
| + | [[File:08.10.2012 carad notI mutagenesis.jpg|500px|thumb|Strong amplification at just above 3000 base pairs from the CarAd PCR using mutagenic primers. Since PSB1C3 is 2070 base pairs and CarAd is 989 base pairs the amplification should run at about 3059 base pairs. 3 different amounts of template DNA were tested in lanes 2-7 as indicated and a positive control showed weaker amplification in lanes 8-9 at about 4000 base pairs.|center]] | ||
| + | <html> | ||
| + | |||
| + | |||
| + | <h2>Week 16 (August 13 - August 17)</h2> | ||
| + | |||
| + | <h3>Desulfurization</h3> | ||
| + | |||
| + | <p>Following on from the progress in the previous week of the mutagenesis, <i>dszB</i>(PstI mutated)-Y63F and <i>dszA</i>(PstI1 mutated)-PstI3 mutagenesis were performed. The gel below shows the digest confirmation. | ||
| + | </html> | ||
| + | [[File:Ucalgary2012 15.08.2012 dszAPstI1&3 dszB Psti y63f muta diges-1.jpg|thumb|500px|center]]<html> | ||
| + | </html> | ||
| + | Another approach was tried to do mutagenesis with a quicker pace. In this approach, after the PCR mutagenesis, the PCR products were purified. Afterwards they were incubated with T4 PNK and ligase. After heat inactivating ligase and T4 PNK, the products were DpnI digested. Subsequently another round of DNA purification was performed. However, the results were unsatisfactory after the digest confirmation. | ||
| + | Sequencing results came back. <i>dszA</i> (PstI1 and PstI3 mutated) and <i>dszB</i>(PstI and Y63F mutated) were good. However <i>dszC</i> (PstI1 and PstI2 mutated) had an insertion next to the PstI1 cut site. Mutagenesis was repeated on the <i>dszC</i>(PstI2 mutated). | ||
| + | <i>dszB</i>(PstI and Y63F mutated)-NotI and <i>dszA</i>(PstI1 and PstI3 mutated)-PstI4 mutagenesis were performed. <b>The gel below shows the digest confirmation</b>. To investigate the actual desulfurisation capability of our initial <i>Rhodococcus</i> source culture for the <i>dsz</i> operon, a desulfurization assay was prepared by innoculating various treatments of the M9 media prepared previously. Also, a conditioning agent composed of 100ml of 95% ethanol, 50ml glycerol, 30ml of 12M HCl (aq) and 70g of NaCl(s) was prepared. The assay relies on the turbidity of a sample containing sulphate ions which are precipitated (hence the turbidometric nature of the assay) upon adding BaCl2(s). Elsewhere, B0034 was constructed with <i>dszC</i> and of the colonies that grew on the plate of the transformed products, colony PCRs were performed and the gel image gave an indication of it being successful. However, further analysis through sequencing showed that the constructions were not successful, and so the constructions were restarted.</p> | ||
| + | |||
| + | |||
| + | <h3>Denitrogenation</h3> | ||
| + | |||
| + | <p>The sequencing for the CarAc/B0034 construct came back with only CarAc and no evidence of a B0034 site preceding it. The confirmation PCR that was run last week using the B0034 forward primer seemed to indicate that it was inserted, however since B0034 is only 12 base pairs the primer may not have been specific enough. The construction on this part was restarted this week. CarAa colony PCR on last week's transformation showed two positive colonies, however upon culturing, miniprepping, and digesting these colonies the plasmid appeared to be too large (3000 base pairs as opposed to the expected 2000). This may be due to a problem of a mislabeled plasmid tube that has been experienced by other team members, causing the gene to be inserted into PSB1AC3 rather than PSB1C3. To test this, a streak plate was made from these colonies and grown on an ampicillin resistant plate. Mutagenized CarAd was successfully transformed into top 10 E. coli cells, cultured, and miniprepped. The miniprep product was digested with NotI to test whether the NotI site was successfully removed. These results were good, as the plasmid ran at 2000 base pairs and the insert at just under 1000 base pairs in the mutagenized CarAd whereas the non-mutagenized CarAd had its insert cut into 2 bands. This will be sent for sequencing next week to confirm the mutation. Unfortunately the sequencing results came back negative for the Ant operon meaning we will need to restart genomic PCR on it next week. Multiple transformations and confirmation PCRs were run on AmdA this week, however none displayed a band size of 1700-1800 base pairs as expected. We have continued to try different digested plasmids as other team members also had difficulties this week with ligation into PSB1C3, possibly indicating a problem with our antarctic phosphatase stock.</p> | ||
| + | <p>The other news from this week was the final results from our carbazole degradation experiment attempting to demonstrate LD2's ability to degrade carbazole and hopefully other nitrogen containing compounds. Unfortunately, these results were very difficult to make sense of as the LD2 that were cultured with carbazole grew, but did not show elevated ammonia levels which would be indicative of carbazole degradation. One of our controls was LD2 cultured with ammonium chloride to test whether LD2 was capable of using ammonia as a nitrogen source. This would pose a problem for us as we were planning to use an increase in ammonia levels as a proxy for carbazole degradation rather than measuring carbazole degradation directly using GC-MS. However, the LD2 showed significantly less ammonia after 44 hours when compared to 26 hours indicating that it may be degraded, either by the LD2 or by an unaccounted for abiotic process. This has lead us to decide to use GC-MS and directly measure carbazole levels in our sample the next time we do this experiment.</p> | ||
| + | |||
| + | |||
| + | <h3>Decarboxylation</h3> | ||
| + | |||
| + | <p>This week, the attempted PstI mutagenesis of CAR was run on a gel. Unfortunately, amplification was unsuccessful, but we had an idea for why this might have occurred. It was discovered that our plasmid of 9kb would need a 7.5 minute extention time, rather than the 1 minute extention time that we had originally programmed into the thermocycler. The mutagenesis experiment was repeated, this time with a 7.5 minute extention time. When these results were analyzed with gel electrophoresis, we had bands appearing to indicate success with the 50ng samples of CAR that we had run, as shown below.</p> | ||
| + | |||
| + | |||
| + | </html>[[File:Band_with_CAR_success_50ng.JPG|500px|thumb|This gel shows 2 bands of CAR that indicate successful mutagenesis of the first PstI site in CAR. The CAR gene is contained in the PET vector. Lane 1 contains the 1 kb plus ladder, while lanes 2,3 contained samples in which 5ng of original CAR+PET plasmid were used, lanes 4,5 contained samples in which 20ng of original plasmid were used, and lanes 6,7 contained 50ng of plasmid sample and underwent mutagenesis by PCR amplification successfully. The bands are slightly over the 9kb band of the ladder, at about 10kb, which is roughly the size of the 10,090bp plasmid. |center]]<html> | ||
| + | |||
| + | |||
| + | |||
| + | <p>After combining these two PCR tubes and incubating with DpnI, we performed a transformation with this DNA and allowed the plate to incubate overnight. Also, we continued to try and verify NPT, but were once again unsuccessful. When we viewed the plate that we had transformed the following day, it was found that we did not have any colonies. We presumed this was because the 9kb plasmid was very difficult to uptake with its large size. Several more transformations were attempted. We also attempted a new procedure called site-directed mutagenesis, which would use a single mutagenesis primer per site, rather than one for each strand, and supposedly had the capacity to mutagenize multiple illegal cut sites at once. We would try to mutate 3 sites with this protocol: the 2 PstI sites, and 1 EcorI site. The PCR was run overnight, with 8 different samples, at two different concentrations of DNA (50ng and 80ng), with four different annealing temperatures. The following day, each sample was incubated with DpnI for 6 hours, and then each was tranformed on a kan plate. A restriction digest of the NPT plasmid was once again performed, cutting with both (E/P) and (X/P) as before, this time using more DNA. An overnight culture of a single potentially successful NPT colony was made, to perform a restrictin digest to see if NPT was present - this would be done over the weekend. Finally, a transformation of the single mutagenesis KAPA PCR of PstI was performed, to hopefully yield more colonies.</p> | ||
| + | |||
| + | |||
| + | <h2>Week 17 (August 20 - August 24)</h2> | ||
| + | |||
| + | <h3>Decarboxylation</h3> | ||
| + | |||
| + | <p>This week, we carried forth with the second and third mutagenesis sites of the CAR gene. Our new plasmid with our first PstI site was PCR amplified with the Kapa system and our EcorI mutagenesis primers to carry out the second mutation, to remove the EcorI illegal cut site from our gene. After the PCR was complete, we ran our product on a gel to see if the amplification worked. Bands indicated success, as shown in the gel below. Following this verification, we mixed our two seemingly successful PCR tubes with 1 uL of DpnI, and left them to incubate for one hour. The PCR product was then transformed into highly competent E.coli cells, and the plates were left overnight in the 37 degree incubator. Unfortunately a mistake prevented colonies from growing, but the process was repeated and our second attempt yielded many colonies. We made overnight cultures of 8 of these colonies, with 4 from each plate, with Kan as the antibiotic for selective growth. The next day, a miniprep was performed on each overnight culture to isolate the CAR+PET plasmids that had hopefully undergone their second mutation, of their single EcorI site. Once the plasmid isolation was complete, we performed a restriction digest using EcorI to cut, in order to verify that the illegal cut site had been removed and the plasmid could no longer be digested with this enzyme. Our success is demonstrated in the gel below. We immediately proceeded with the third mutagenesis, which would this time be performed on the second (and last) PstI site. Using our new plasmid now with two illegal cut sites removed, we performed an amplification PCR, using our PstI(2) primers and the Kapa system. After running this on a gel, we determined bands of success, as shown below. We then once again treated our PCR amplified tubes with DpnI for one hour, and then transformed into highly competent E.coli. The plates were left overnight to allow colony growth. Our second accomplishment of the week was successfully amplifying the OleT gene from Jeotgalicoccus, by using the Kapa system. This was very successful, as shown in the gel below. We then performed the cycle-pure spin protocol to remove contamination from our PCR product, and then proceeded to perform a restriction digest on the product with HindIII and AscI so that we could ligate it into the PET vector. After this ligation was complete, it was transformed with E.coli on a Kan plate, and left overnight to incubate. Unfortunately we do not see colonies yet, but the incubation is not complete yet, and might still be successful. Thirdly, we obtained the PetroBrick stock from the Washington team on Tuesday, and immediately proceeded to transform it. We obtained a great deal of colonies, and then performed colony PCR on 12 of these colonies to ensure that our ~2400bp PetroBrick was present in these cells. Last but not least, we have still been working on a successful purification of NPT from a transformed plate. A colony PCR indicated that NPT was potentially in a few of these colonies, so we made overnight cultures of these four colonies, and performed a miniprep the next morning, and then a restriction digest to verify that the gene was present.</p> | ||
| + | |||
| + | <h3>Denitrogenation</h3> | ||
| + | |||
| + | <p>The CarAa was re-streaked onto ampicillin plates and grew, probably showing that it was ligated into PSB1AC3, not PSB1C3, which explained the irregularity of our confirmation digest last week. It was sent for sequencing, but in case it was incorrect we also started a new CarAa pipeline by PCRing it from the Pseudomonas genome again and redigesting and ligating. It was transformed into PSB1C3 over the weekend. CarAc did not see a lot of progress this week as we spent much of the time attempting to construct a RBS (B0034) in front of it, but were unsuccessful. CarAd was also subjected to this construction with B0034 and we were able to send it for sequencing by the end of the week. Multiple new constructs were also started for CarAd in case the sequencing is negative. The Ant operon was successfully transformed and confirmed by colony PCR this week, after which it was grown up and sent for sequencing. We also began to attempt mutagenesis on one of the EcoRI sites in the operon, but had no amplification of the plasmid. We also re-digested some PCR purified product to start another ligation in case sequencing failed. After many attempts we were also able to finally successfully transform an AmdA/PSB1C3 plasmid into top 10 cells this week. After a miniprep they showed bands of about 1800 base pairs on the confirmation digest gel, so they were also sent for sequencing this week and had mutagenesis started on the illegal SpeI site. Unfortunately, the sequencing sent early in the week came back with ambiguous results. The forward reaction for AntABC was a match, but its reverse reaction matched to CarAa. Both CarAa reactions matched nothing. Since these two genes are over 20 000 base pairs apart on the template plasmid it is virtually impossible that they were actually combined into one plasmid (PCR extension time would not have been long enough and even if it was the product would have run much above the 3000 base pair mark). This is likely a mistake on Eurofins end, so we are not 100% sure if we have 1 good gene, both of them, or neither. Also the sequence for mutagenized CarAd showed only about the first 100 base pairs, which were correct however not long enough meaning it was a poor reaction and inconclusive. Finally, we also re-started our carbazole degradation assay using the GC-MS at biological sciences. This process will take 2 weeks to demonstrate LD2's ability to degrade carbazole. We are forcing it to use carbazole as a carbon and a nitrogen source in one sample and as only a nitrogen source in another sample (this sample has had glucose added to it).</p> | ||
| + | |||
| + | <h3>Desulfurization</h3> | ||
| + | |||
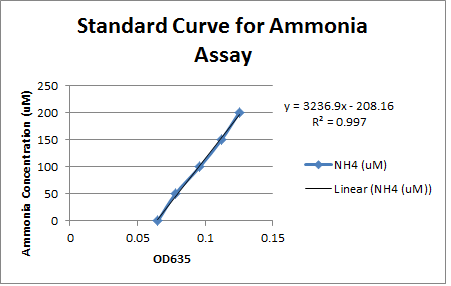
| + | <p>This week, progress was made in determining the desulfurization activity of our <i>Rhodococcus</i> strain as measured by the sulfate release using a turbidometric assay. WE encountered numerous challenges in our prescribed protocol as the concentrations that we are using to prepare out standard curve may be too dilute, or the composition of out conditioning agent may be flawed. Additionally, steps were taken to determine the decomposition of DBT to 2-HBP through GCMS analysis, but due to a preparation error, the DBT was added to a growth solution of M9 media prematurely and the autoclaving process decomposed the DBT releasing a yellow colouration into the solution. These two approaches in determining the desulfurization capability of the dsz operon will be further investigated. </p> | ||
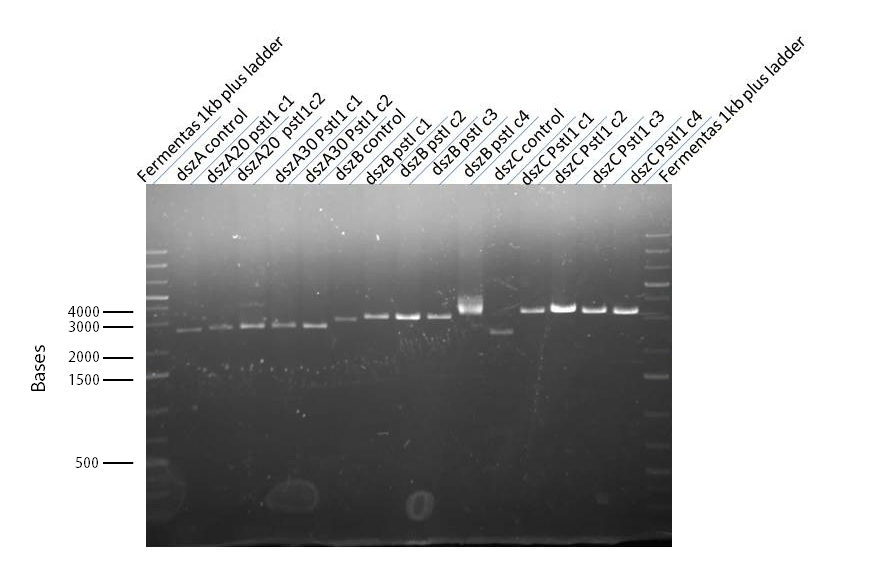
| + | |||
| + | <p>Since the dszC second mutagenesis had proven to be unsuccessful last week, the <i>dszC</i>(PstI2 mutated)PstI1 mutagenesis was repeated. Also <i>dszA</i>(PstI1,3,4 mutated) PstI2 mutagenesis was performed. <i>dszA</i> and <i>dszC</i> were sent for sequencing on Wednesday. <i>dszB</i> was sent for sequencing on Friday. Sequencing results of <i>dszA</i> and <i>dszC</i> were back by Friday. <i>dszC</i> was successful. However, <i>dszA</i> contained an insertion next to the binding site of PstI4 cut sit, so the last two mutations must be redone. <i>dszB</i>(PstI and Y63F mutated)-NotI-mutagenesis was also repeated in case the result of the sequencing was not successful. These constructions were repeated. J13002-<i>dszB</i> and B0034-<i>dszC</i> constructions were attempted, however they were not successful as indicated by cPCR. Constructions of J13002/<i>hpaC</i> were carried out and also came back negative in sequencing, however B0034/<i>katG-LAA</i> was sequence confirmed.</p> | ||
| + | |||
| + | <h2>Week 18 (August 27 - August 31)</h2> | ||
| + | |||
| + | <h3> Decarboxylation </h3> | ||
| + | |||
| + | <p>This week, the primary objectives from last week were continued. Another restriction digest of OleT and then a ligation was performed, in order to see if we could obtain more colonies following transformation. After preparing overnight cultures and minipreps of eight of these colonies, we attempted to cut the OleT/PET vector with EcorI to verify that the gene had successfully inserted into the vector. Unfortunately, we did not have any luck, even after several attempts. Our attempts to verify this will continue. We also performed ligations with NPT (cut with XbaI and PstI) with JI3002 (cut with SpeI and PstI), so that we could produce a biobrick with JI3002 before NPT to act as a promoter and ribosome binding site. We decided on how we will build our two primary constructions, and where each part will be. Also, we attempted to amplify the CAR gene in the PET vector (with 3 mut sites replaced) with biobrick primers, so that we could get it inserted into a biobrick plasmid. After several tries, this amplification was successful.</p> | ||
| + | |||
| + | <h3> Desulfurization </h3> | ||
| + | <p><i>dszB</i> sequencing results came back as successful. <i>dszA</i>(PstI1,3 mutated)-PstI2-mutagenesis was performed and sent for sequencing. Also <i>dszA</i>(PstI1,2,3 mutated)-PstI4-mutagenesis was performed, and this was also sent for sequencing. </p> | ||
| + | <p> Constructions of J04500 with <i>hpaC</i>, <i>dszB</i>, and <i>katG-LAA</i> were performed. Transformations were carried out at the end of the week. | ||
| + | |||
| + | <h3>Denitrogenation</h3> | ||
| + | |||
| + | <p>The new stock of CarAa that we had PCRed from LD2 was sent for sequencing this week and came back as a match, so we have begun constructing a RBS site and a TetR promoter (J13002) in front of it. At this point all the Car genes have been successfully amplified, inserted into a biobrick plasmid, and sequence verified. We also continued to attempt to construct CarAc with an RBS site (B0034) in front of it, but had no success. On a better note, we confirmed this week that CarAd was mutated properly and we were also able to insert an RBS site in front of it. The next step for this gene will be to attempt a plasmid switch as it is currently in PSB1A3 and needs to be in PSB1C3 for submission to the parts registry. After sending a new batch of the Ant operon for sequencing we got the same confusing results as we did last week. We therefore hypothesized that our primers were annealing to a homologous region that is similar to the Ant genes and very close to CarAa. This means that we need to restart PCR on this gene as all of our current product is incorrect. The sequencing results for AmdA that came back this week were also somewhat confusing as they did not quite match the sequence we expected. However, they were a 100% match for the AmdA gene in a different strain of Rhodococcus erythropolis. This means that DSMZ probably sent us the wrong strain of bacteria and that we were very lucky that the primers we designed based off of the expected template actually worked to amplify a slightly different gene in another strain. The good news is that this new sequence contains no illegal cut sites, which explains why mutagenesis was not working (there was nothing to mutate) and allows us to immediately begin construction of J13002 in front of AmdA this week. </p> | ||
| + | |||
| + | <h2>Week 19 (September 3- September 7)</h2> | ||
| + | |||
| + | <h3> Denitrogenation </h3> | ||
| + | |||
| + | <p>CarAd/B0034 was successfully plasmid switched into PSB1C3, making it ready for submission. Construction of J13002 and CarAa may have been successful as the digest of the miniprep looked promising. It will be sent for sequencing next week. Unfortunately there was no progress made on the constructions of AmdA with J13002 or CarAc with B0034. These will be redigested and religated for next week. We could not amplify the Ant operon despite trying many variations of PCR using high fidelity KAPA polymerase. This is probably due to the multiple, large regions of homology on the operon that the reverse primer binds to making amplification difficult. Because the function of this gene can theoretically be replaced by AmdA, we may scale back efforts dedicated to it, as the chances of successful amplification appear slim. </p> | ||
| + | |||
| + | <h2>Week 20 (September 10 - September 14)</h2> | ||
| + | |||
| + | <h3> Denitrogenation </h3> | ||
| + | |||
| + | <p>Constructs that continued this week included AmdA with J13002, CarAa with J13002, and CarAc with B0034. Some sequencing was sent, but no positive results were obtained. A new approach to biobrick the Ant operon was implemented this week. A new forward primer was designed that would anneal about 1500 base pairs upstream of the Ant operon, avoiding the region of homology that is repeated at various points on the plasmid. Once this region is successfully amplified we could use this amplified DNA as a template for another PCR reaction which would use the normal Ant primers. Since the template would no longer include regions of homology these primers should be free to work properly and amplify the Ant operon. The PCR using the new primers was started this week, and although amplification at 4500 base pairs was observed as expected, there was also a lot of non-specific amplification. This means that to purify the amplified DNA that we want and avoid the other products we will have to perform a dreaded gel extraction next week. </p> | ||
| + | |||
| + | |||
| + | |||
| + | </html> | ||
| + | }} | ||
Latest revision as of 18:44, 21 September 2012


Hello! iGEM Calgary's wiki functions best with Javascript enabled, especially for mobile devices. We recommend that you enable Javascript on your device for the best wiki-viewing experience. Thanks!
Team
THIS PAGE NO LONGER EXISTS AND WILL NOT APPEAR ON THE WIKI. Please migrate your data and information to your respective pages (as outlined in the email I sent you a while back.) -PW
you guys need to update your notebook more?
we working hard, y u get so mad? :'(
Week 1 (May 1-4)
The following section covers wetlab aspect of our overall project focusing on microbial conversion of naphthenic acids into economically-valuable hydrocarbons. The approach taken in this endeavour will be from four strategic starting points - ring cleavage, decarboxylation, denitrificaiton, and desulfurization. Overall, the 'hydrocarbons' aspect of the project is a critical one to our overall design and construction of a biosystem capable of not only detecting but also converting naphthenic acids in what will be an economically viable solution to the remediation and recovery of tailings pond water, while removing these toxic compounds before they can have significant detrimental impact on the environment.
Week 2 (May 7-11)
Decarboxylation
For the decarboxylation sub-project, the second week was entirely focused on literature research and the practice of basic laboratory techniques. 8 potential pathways were identified as potential candidates for naphthenic acid decarboxylation. The first of these would utilize only the University of Washington's "PetroBrick" (from iGEM 2011), consisting of the genes encoding the enzymes acyl-ACP reductase (AAR) and aldehyde decarbonylase (ADC). We have planned to verify the PetroBrick in the distribution plates and test its efficacy on naphthenic acids in the coming weeks. If this proves to be unsuccessful, we will begin investigating the alternative approaches, beginning with replacing AAR with carboxylic acid reductase (CAR) from Nocardia iowensis, a very unspecific reductase shown to work on structures resembling naphthenic acids. Failing this, the remaining pathways will be examined; however, the disadvantage in these pathways is their direct reliance on the success of the other steps, as the naphthenic acids must be degraded to the point of resembling branched-chain fatty acids (since all remaining pathways are related to fatty acid metabolism).
Denitrification
In the first two weeks of iGEM our group has focused on reviewing literature regarding the bioremediation of nitrogen groups attached to naphthenic acids. The most prevalent N heterocycle is carbazole, representing 75% of total nitrogen by mass. The upper pathway of carbazole biodegradation is catalyzed by the enzymes coded for by the car operon, CarA (CarAaAbAd), CarB (CarBaBb), and CarC. These enzymes convert carbazole to anthralinic acid. The lower pathway is catalyzed by the enzymes of the ant operon, antA, B, and C, yielding cathecol while releasing CO2 and NH3. The car and ant operons are both regulated by the Pant regulator which is induced by the protein, antR. CarAa also has its own promoter which is not induced by antR. We have also investigated an alternative pathway using CarA combined with an amidase (amdA) that selectively cleaves NH2 from an intermediate of the car pathway. This could bypass much of the car/ant pathway and is possibly more efficient.
We have decided to use Pseudomonas resinovorans and Rhodococcus erythropolis to amplify these genes from. CarABC and AntABC from P. resinovorans has been shown to have a wide range of nitrogen containing substrate specificity. R. erythropolis contains the amdA gene that we wish to use, and some evidence suggests that it may also be able to degrade sulfur rings through its CarABC pathway.
In addition to our research we have also been learning some of the lab techniques we will be using this summer. This includes transforming a plasmid into E. coli, plating and selecting for bacteria containing the plasmid, verifying with colony PCR, performing a mini-prep and a restriction digest.
Desulfurization
Along with the rest of the team, this week was dedicated to familiarizing ourselves on the protocols that will be utilized during this years project; specifically the polymerase chain reaction, gel verification, preparation of overnight cultures, as well as developing a procedural flowchart to transform competent cells with registry biobricks. With regards to our sub-group specific goals, we reviewed the current available literature around various industrial and laboratory approaches to desulfurization of organic groups, especially in the petroleum industry. This included a comparison of non-biological processes such as conventional hydrodesulfurization, which is currently employed in petroleum product refinery stages, and how a biological approach would supplement and perhaps even offer several advantages over these methods. Current limitations to biological desulfurization, however, include such factors as biocatalyst stability, enzyme specificity, desulfurization rate, and a need for a carbon source to regenerate co-factors. We also identified the enzyme desulfinase (DszB) as being one of the bottlenecks in the desulfurization 4S pathway. Overall, our goals moving forward involve determining the specific pathways involved in the desulfurization process as well as the reaction conditions we would want to employ, and identifying specific model compounds in addition to dibenzothiophene (DBT) that we could use to test the effectivity of our biosystem in order to determine its functionality in the conversion of naphthenic acids to economically valuable hydrocarbons.
Ring Cleavage
This week we mainly researched aromatic ring cleaving using intra- and extradiol dioxygenases from species of Pseudomonas and Bacillus but also began literature searches on aliphatic ring cleavage done by monooxygenases.
Week 3 (May 14-18)
Decarboxylation
The third week included some additional literature investigation in the first two days. The iGEM distributions arrived this week, and verification began on May 17th by transformation into E. coli, followed by colony PCR on May 18th (using standard protocols on the wiki). Additionally, primers were designed for CAR in N. iowensis, along with primers for Nocardia posphopantetheine transferase (NPT), a second enzyme required for optimal function in the former, and a short list of contacts were acquired to request the donation of the required strain (called NRRL 5646) from researchers who have worked with it previously. A sort of form email was drafted for this purpose, and should this be unsuccessful, we will be purchasing the strain from DSMZ (http://www.dsmz.de). In the following week, we will begin with development of overnight cultures and gel preparation.
Ring Cleavage
In the second week we continued to do more research on the degradation of alicyclic compounds and found two strains of bacteria that contained genes needed for this process. The first strain, Thauera butanivorans, contains the genes required to activate the ring by adding a hydroxyl group. The gene is called butane monooxygenase and is composed of three subunits, a hydroxylase, a reductase, and a regulatory component. The second strain, Acinetobacter sp. SE19, contains the genes needed to oxidize the alcohol, formed by butane monooxygenase, and to cleave the ring. There is a cluster of nine genes that perform this oxidation and cleavage but only six are involved directly.
Desulfurization
Building on the previous week's literature review, the 4S pathway was recognized as the preferred biological mechanism that we would explore in devising a desulfurization biosystem. Of specific interest is the dsz operon consisting of the genes for dszA, dszB, and dszC which selectively and non-destructively remove the sulfur from the hydrocarbon structure, and therefore preserves the carbon skeleton. In addition to these, another dsz gene exists. dszD, which codes for a FMN:NADH reductase, is an essential component of the pathway, but not part of the operon. Instead, it is encoded on the chromosome. The enzyme produced by this gene is required to regenerate the FMNH2 consumed by the reactions carried out by DszA and DszC. Rhodococcus erythropolis IGTS8 is the most studied model organism in investigations of the 4S pathway, and has been shown in many different research endeavors to be capable of converting DBT to 2-HBP.
INSERT PATHWAY DIAGRAM
An alternative to the DszD gene is HpaC, an oxidoreductase encoded in the E. coli W genome. This enzyme has been shown to increase the rate of desulfurization by x amount find citation Following this, other protocols added to our growing lab methods 'toolkit' were a restriction digest protocol, PCR purification, and finally, DNA construction digest. Aims moving forward include obtaining strains of the R. erythropolis , while also executing a timeline devised to biobrick, test, and incorporate the genes necessary in the above processes in a biobrick circuit.
Week 4 (May 22-25)
Decarboxylation
This week, a gel was prepared for the colony PCR prepared last week, and the gel was run, yielding results that were very difficult to see. The PCR and gel electrophoresis process were repeated, yielding the following gel:
Results indicate successful transformation with the PetroBrick. Lane 1 contains the 1 kb plus ladder, while Lanes 2-11 contain the results of colony PCR. Each of these bands correspond to the PetroBrick vector, which is 2070bp, indicating that the transformation was successful. Lane 12 contains a positive control, which varies distinctly from the colony PCR results. The negative control lane is empty as expected.
Overnight cultures were grown from the colonies were prepared this week. Sigma compounds - for the purpose of testing the PetroBrick on naphthenic acid analogues - were selected from the list provided. The compounds to be used include cyclohexanepentanoic acid, cycohexane-1,1-dicarboxylic acid, and benzo[b]thiophene-3-acetic acid. A mini-prep was completed on the overnight cultures prepared earlier in the week, according to protocol obtained from the wiki. The DNA concentration in the resulting samples was measured by nanodrop to confirm successful plasmid extraction, and the resulting DNA concentrations were as follows:
- Tube 1: 44.9 ng/mL
- Tube 2: 54.2 ng/mL
- Tube 3: 54.4 ng/mL
- Tube 4: 41.6 ng/mL
- Tube 5: 34.3 ng/mL
Because tube 3 (prepared from Colony 3, Plate 3) had the highest concentration, it was to be used for the eventual sequencing of the plasmid. The restriction digest was completed also, but it was not run on a gel this week; this was left for Week 5.
Contents |
Desulfurization
This week was kicked off with a project development meeting with Emily and David, and we devised a protocol for biobricking the hpaC gene. Additionally, methods to place the genes coding for the 4 enzymes, DszA,B,C and HpaC into a single construct were explored. Within the lab, the PCR performed on the resuspended pUC18-hpaC was not successful initially. Furthermore, we ordered the substrates/compounds that we intend to use for desulfurization tests. Once the substrates and the Rhodococcus strain arrive we are going to test how effectively the bacteria can desulfurize different sulphur-containing compounds that resemble naphthenic acids. Finally, we came across a paper published by REFERENCE , whos team had developed an improved efficiency DszB through site-directed mutagenesis in 2007. This was through a point mutation to the gene, converting a tyrosine at position 63 to a phenylalanine residue. A member of this team was contacted to request the plasmid that contains the mutated gene. The conversion step carried out by DszB is the major bottleneck in the 4S pathway and if a strain or sample containing this mutation was obtained, it would significantly bolster our later testing efforts on DBT, as well as other compounds such as thiophane.
Denitrification
This week we reviewed the primers listed in the database and also designed some new ones. Primers for CarAa, CarAc, and CarAd were designed individually, while primers for CarBaBbC and AntABC were designed to encompass multiple genes in a sequence. These primers were designed to be used on Pseudomonas putida which we decided to use as our gene source since it was available to us as opposed to ordering Pseudomonas resinovorans. We also designed a primer for the AmdA gene from Rhodococcus erthyroplois. In addition to ordering these primers we also ordered the nitrogen containing compounds that we will need to test these enzymes on. We decided on using carbazole to make sure the enzymes can perform their natural function as well as pyrrolidine to test them on a similar ring structure. We also ordered cyclohexamine in order to independently test the function of the alternative AmdA pathway. Finally, we decided to eventually order 4-Piperidine butyric acid hydrochloride to test how the enzymes will work on nitrogen containing naphthenic acids. However, we decided since it is a very expensive compound we would wait to make sure the enzyme's work on more simple compounds before ordering it.
We also started our work in the wet lab by plating colonies and making an overnight culture of -80 freezer stock Psuedomonas putida on Thursday. We also resuspended the primers for CarAa, CarAc, CarAd, AntA, AntB, and Ant C that were already in the database. We were able to use the colonies that grew on the streak plates to start a colony PCR to attempt to isolate each of these genes on Friday.
Ring Cleavage
We started looking into more organisms that can carry out cyclohexane degradation and found one called Brachymonas petroleovorans. This organism is capable of both hydroxylating cyclohexane and carrying out cyclohexanol oxidation. We are also hoping to find evidence that Pseudomonas fluorescens, Pf-5 has genes that can cleave aliphatic rings, however, first we have to determine if Pf-5 can grow in the presence of butylcyclohexane. To do so we carried out an assay, using LB, testing the toxicity of butylcyclohexane on Pf-5. After 24 hours of incubation we took OD600 readings¬ which suggested that the presence of butylcyclohexane did not have effect on cell growth. We also looked up Pseudomonas fluorescens Pf-5 in the Pseudomonas Genome Database and found genes that code for proteins with similar functions to what we have found for aromatic and aliphatic ring cleavage.
Bioinformatics/Modelling
Handy bioinformatics tools
FMM (From Metabolite to Metabolite) is a critical tool for synthetic biology. FMM can reconstruct metabolic pathways form one metabolite to the other one. <a href:http://fmm.mbc.nctu.edu.tw/>http://fmm.mbc.nctu.edu.tw/</a>
This webpage runs the software that performs the enumeration of all pathways for target compounds in KEGG. More precisely, the desired KEGG compound is submitted in order to get the map of all pathways connecting the compound to metabolites endogenous to E. coli. Optionally, the full map containing the supplements can also be requested. <a href:http://bioretrosynth.issb.genopole.fr/tools/metahype/>http://bioretrosynth.issb.genopole.fr/tools/metahype/</a>
DNA Designer 2.0 is a Drag and drop standalone software, runs on Windos, MacOS. <a href:https://www.dna20.com/secure/order.php?page=genedesigner2>https://www.dna20.com/secure/order.php?page=genedesigner2 </a>
DNADesign is a Web-based application <a href:http://genedesign.thruhere.net/gd/> http://genedesign.thruhere.net/gd/</a>
Modelling
Constraint-based flux analysis - modeling microorganism metabolic network and perform flux analysis.
The metabolic network is translated in to a matrix and then the simulation will calculate a pathway that optimizes the goal (max biomass or max production).
The network will be reconstructed (gene/reaction can be added or deleted) to enhance the expected pathway to degrade target compounds. The media (compounds, thermo-conditions) will be manipulated to find the best condition(s) that satisfy the microorganisms to digest the target compounds and reach a good growth rate (say above threshold).
Week 5 (May 28 - June 1)
Decarboxylation
On Tuesday (May 29), a gel was run on the restriction digests of the extracted plasmids from Week 4, which appeared to confirm the successful transformation of the PetroBrick, showing clear bands at the pertinent locations. The gel is as follows:
Lane 1 contained the 1 kb plus ladder, while lanes 2-6 contained the restriction digest results, which had used the five different miniprep tubes with plasmids isolated from five different colonies. As can be observed in the gel, the upper row of bands corresponds to 2392bp, indicating the petrobrick part. The lower set of bands corresponds to the PetroBrick pSB1C3 vector at 2070 bp. These results suggest that the PetroBrick plasmid had been successfully purified. The black circle indicates undigested plasmid in lane 3. There simply means that not all of the plasmid was digested into two separate DNA strands. It indicates the entire size of our PetroBrick plasmid.
Based on the apparent success of the gel, the products were sent away for sequencing. Long-term stocks were prepared for the alkane production medium outlined in University of Washington's PetroBrick protocols from 2011 (see https://2011.igem.org/Team:Washington/alkanebiosynthesis). These stocks are as follows:
- 1 L of 1M Tris (pH = 7.25)
- 10 mL of 1 mg/mL Thiamine
- 10 mL of 10% Triton x-100
- 1 M MgSO4
- 0.1 M FeCl3 (anhydrous)
The medium itself is to be prepared in Week 6 once the results of sequencing are (hopefully) acquired.
Ring Cleavage
This week we redid the previous assay with Pf-5 and butylcyclohexane using glass vials instead of falcon tubes. These vials allowed for more surface and better containment of the volatile compound. We carried out this assay using both LB and M9-MM. After 24 hours we took OD600 readings and obtained similar results as last week. The growth was considerably decreased in the M9-MM, however we are not sure whether the bacteria were able to use the compound as a carbon source because the MM contained glucose. The fact that the growth decreased showed that they might depend on the glucose in the media to grow.
Denitrification
On Monday and Tuesday we used the database primers to attempt to isolate CarAa, CarAc, CarAd, AntA, AntB, and AntC from the Psuedomonas putida we plated last week. CarAd, AntB, and AntC were all put in the gradient PCR machine to account for the wide range of their primers’ melting points, while the others were done via regular PCR. Unfortunately, no positive results were obtained from these reactions. The PCR on CarAc and AntA was repeated on Wednesday, still not giving positive results. Finally, we attempted to use a salt concentration gradient on the PCR reaction for CarAc and AntA, using concentrations that ranged from 1.0 microlitres/tube to 2.0 microlitres/tube in increments of 0.2 microlitres/tube. This helped as CarAc showed bands in samples that had concentrations of 1.2 and 1.4 microlitres/tube. AntA also showed bands, however they were not the correct size, indicating contamination and/or non-specific annealing of the primer. The positive control also showed bands of the correct size. Earlier in the week we also made overnight cultures of 3 environmental strains (28, 29, 30) of Psuedomonas putida from -80 glycerol stock. On Friday we performed a genomic prep on these cultures, and plan on attempting to amplify genes from the isolated DNA next week.
Desulfurization
Since we wanted to make sure we would not run out of pUC18(plasmid containing the hpaC gene), we transformed some E.coli cells with it. We grew them on plates containing A, K, T and C antibiotics and they only grew on A. Therefore pUC18 has A resistance. We did a three sets of PCR with primers designed against hpaC, one using 1/10 dilution of pUC18, the other using 1/100 dilution of pUC18 and one with the colonies we had just obtained by transforming the E.coli cells. The PCR worked and we saw bands of the same size for all three sets of PCR. (Unfortunately, the picture we saved is not a good one since some of the bands faded away under UV due to prolonged exposure. Following this, PCR purification was performed to obtain the pure hpaC with biobrick prefix and suffix attatched to gene, which would allow us to insert the sequence into a biobrick standard backbone. 3 sets of digestion, ligation, and transformation (using pairs of X&P enzymes, E&S enzymes and E&P enzymes) were carried out in order to insert the hpaC gene into the pSB1C3 vector. All the sets grew successfully. Following the above successes with hpaC, the arrival of our Rhodococcus strain afforded us the opportunity to begin investigation of the Dsz operon using the primers current in our possession. This strain is an environmental isolate that has been shown by someone to be an active desulfurizer. The gram-positive nature of the strain also dictated we explore various lysing strategies before the genes encoding the Dsz enzymes could be amplified for further purification and biobrick construction steps. PCR was carried out using dszA primers on three different treatments {microwave, lysate buffer, and a control} which yielded banding pattern around 1200 base pairs for the lysate treatment (2%SDS and 10% tritonX-100, plus heat for 5mins at 98C). DO WE HAVE A PICTURE
Week 6 (June 4 - June 8)
Decarboxylation
This week, we first made a streak plate of our confirmed Petrobrick E.coli Colony 3, Plate 2, as well as a long-term glycerol stock for storage. In accordance with the Washington team’s protocols, we prepared our M9 minGlucose Media (Production Media) with the following reagents:
- 75mL ddH2O
- 0.6g Na2HPO4
- 0.3g KH2PO4
- 0.05g NaCl (855.6mL 1M solution)
- 0.2g NH4Cl
- 20mL of 1M Tris (pH = 7.25)
- 1mL of 10% Triton
- 100mL of 1mg/mL thiamine hydrochloride
- 10mL of FeCl3 (anhydrous)
- 100mL of MgSO4
**3.0g glucose needed to be added later after the mixture was autoclaved, as glucose is known to caramelize under the conditions of the autoclave.
We also prepared an overnight stock containing 5mL of LB broth inoculated with E.coli from our verified colony. The following day we measured its optical density, demonstrating that our OD was 1.311, suitable for our purposes. We obtained our PetroBrick sequencing results with a complementarity of 87%, as compared to the theoretical PetroBrick in the PartsRegistry, which was deemed by iGEM team leaders to be high enough to continue in our protocol. We also performed glucose filtration, transferring 3g of glucose into our Production Media solution after it had been autoclaved in order to ensure sterile transfer. On the final day of the week, we were able to start our General Production Protocol that would be followed by a 48 hour incubation. It was in this time that hydrocarbon production was reportedly supposed to occur. Use of the GC would occur in the following week to test for alkane production.
Ring Cleavage
This week we made subcultures from Pf-5 in M9-MM into M9-MM without glucose to avoid carryover of glucose, to insure that the bacteria use butylcyclohexane as their only carbon source. We also took GC readings and compared experimental values to standards made with water and butylcyclohexane (butCH). Readings showed a decrease in amount of butCH in the headspace, compared to standards, indicating that the compound was being degraded by Pf-5. Then, we made subcultures from Pf-5 in M9-MM without glucose into the same media and butCH to decrease carry over of glucose and did time zero readings. Furthermore, we made subcultures from original Pf-5 in M9-MM without glucose into the same media and toluene to test for pathways that degrade aromatic structures. To identify possible genes for the pathways we started running blasts on the Pf-5 genome looking for genes that are homologous to ones that we have been looking at for the two ring cleavage pathways (aromatic and aliphatic). In addition we started a verification of XylE (aromatic ring cleavage enzyme) by transforming part K118021 (XylE with a Pcst promoter) from the parts registry and did two colony PCRs. unfortunately none of them showed successful results and only the positive control showed an appropriate length DNA.
Denitrification
We started off this week by determining the DNA concentration of our genomic prep samples from last week using the nanodrop. DNA concentration for all three putida strains was at least 1000 ng/microlitre, well above what was needed for PCR. 1/2 and 1/3 dilutions were prepared for all three strains so as not to have an excess of template DNA in PCR reactions. PCR was performed on all 3 strains using primers for CarAc, CarAd, AntA, AntB, and AntC using 6 replicates per gene. The only successful amplification appeared to be AntB and CarAc, both from strain 28 (with weaker bands in strain 29). We then performed another PCR, just on those two genes with an increased amount of Taq polymerase to hopefully get enough amplified DNA to move forward with. This resulted in strong bands for both at the expected size. We then performed PCR purification using the Qiaquick kit and obtained samples containing 33.5 ng/microlitre of AntB DNA and 129 ng/microlitre of CarAc DNA. These concentrations were both sufficient to begin a restriction digest and ligation of these parts into vector PSB1C3. Next week we hope to verify the results of the restriction digest, continue to amplify CarAc and AntB from strain 28, and hopefully submit a biobrick for sequencing.
Desulfurization
In order to confirm the hpaC biobrick construction, two sets of colony PCR were performed, choosing white colonies from the 3 plates we grew last week (white colonies indicate a loss of the RFP generator in the pSB1C3 backbone, and therefore allow for weeding out of the colonies which are simply the original plasmid vector). These reactions were carried out both with hpaC primers and with stanndard biobrick primers designed against the plasmid backbone. After running them on the gel we saw equal bands for the PCR reactions performed using hpaC primers FIRST PIC(However, a PCR using biobrick primers was performed later and the same results were obtained). Colonies 1(-) and 5(-) were used to make overnight cultures, which were then miniprepped the following day to obtain the plasmid DNA of the putative hpaC biobrick. Digestions were performed on the miniprep products using EcoRI and Pst to look for part size as further verification for the genes presence in the plasmid. The results were good and two bands were observed on each column (one for vector and the other for hpaC)(second figure). hpaC was sent in for sequencing.
PCR reagents were prepared to re-test/confirm previous results of dszA amplification following two different lysing treatments (microwave + lysate buffer). This time, all three genes were amplified and gel verification showed clear banding patterns around 500bp range for all three genes for the microwave treatment. WHY DID WE PROCEED WITH THIS THAT ISNT THE RIGHT SIZE Remaining PCR products were run on a gel and extracted for further purification steps; however, presence of any genetic material were not confirmed through nanodropping which raised concerns about the composition of the purified products, the success of the initial amplification step, or perhaps even the lysis treatment. Further experimentation will have to be carried out to troubleshoot.
Week 7 (June 11 - June 15)
Ring Cleavage
Since last week we continued taking GC readings on the Pf-5 incubated with butylcyclohexane and toluene. At this point the Pf-5 had been incubated with butylcyclohexane for 6 days and with toluene for 5 days. The amount of both butylcyclohexane and toluene decreased by a large amount in the experimental cultures but the amount of each in the abiotic standards also largely decreased by about the same amount. This lead us to conclude that the reason for decrease was not due to the bacteria metabolizing the compound, but due to unknown abiotic factors. To measure relatie amounts of bacterial growth we took OD600 readings for both cultures which were very low and indicated that there was little bacterial growth. The verification of XylE from the parts registry was continued using a miniprep on three overnight cultures from part K118021. The concentrations were very high likely due to genomic contamination. A second part (J33204) containing XylE and an rbs site was transformed and a colony PCR was done on part J33204 but the gel did not indicate successful results, again only the positive control worked. No positive colonies were found.
Decarboxylation
After preparing our production media 48 hour incubation sample for gas chromatography and extracting with ethyl acetate, we left our sample for use in the GC. Our results indicated a small hydrocarbon peak, indicating a possibility that hydrocarbon production had been a success, but it was not distinct enough to make this conclusion. The procedure would be repeated, this time creating sigma compound solutions as well. These sigma compounds would resemble naphthenic acids. In order to ensure that hydrocarbon peaks would be produced from these compounds and not from glucose, we prepared a new production media lacking glucose, in hopes that incubated bacteria could survive for long enough without a food source to decarboxylate a sigma compound to some extent. Since the sigma compounds we initially selected for our purposes had not arrived as of yet, we opted for others: Cyclohexanepentanoic acid (CHPA), cyclohexanecarboxylic acid, and 1,4-cyclohexanedicarboxylic acid. We performed 8 overnight cultures and prepared sigma compound solutions. Despite our many attempts, we were only able to acquire an OD of about 0.6 for these 8 tubes. We then performed our production protocol with the tubes, preparing them with the following reagents and inoculating them with the contents of our broth cultures:
- Tube 1: Glucose
- Tube 2: Glucose
- Tube 3: Glucose and CHPA
- Tube 4: Non-glucose and CHPA
- Tube 5: Glucose and cyclohexanecarboxylic acid
- Tube 6: Non-glucose and cyclohexanecarboxylic acid
- Tube 7: Glucose and 1,4-cyclohexanedicarboxylic acid
- Tube 8: Non-glucose and 1,4-cyclohexanedicarboxylic acid
In the above tubes, “glucose” indicated that glucose-containing production media was used, whereas “non-glucose” indicated that the production media lacking glucose was used. In each case where a sigma compound was added, it was present at a concentration of 25mg/L. Each production tube was then left for a 48 hour incubation over the weekend, to perform gas chromatography tests the following week.
Denitrification
The purified PCR products for CarAc and AntB were digested with restriction enzymes EcoRI+SpeI, EcoRI+PstI, and XbaI+PstI. However, only PSB1C3 plasmids that had been digested with EcoRI+SpeI and EcoRI+PstI were available to attempt ligation. Gel results were inconclusive on the restriction digest product as the plasmid size appeared much larger than expected. However, transformation was still attempted on the ligation products for both genes (both using EcoRI+SpeI ligation into PSB1C3 plasmid). The transformation products were plated onto chloro plates to select for colonies that had the chloro resistance gene on the PSB1C3 plasmid. Also this week another round of PCR was performed for the Car and Ant genes, however all, but CarAc showed bands in the negative control lane, possibly indicating contamination or the formation of primer dimers. The CarAc bands were fairly weak and a PCR purification resulted in very low DNA concentration, insufficient to move onto restriction digest. Next week's plan hinges heavily on the result of the transformation from this Friday.
Desulfurisation
This week, we focused on amplifying dsz genes from our Rhodococcus strain for construction into biobricks. We also wanted to purify the PSB1C3-hpaC and pUC18-hpaC plasmids to replenish our current stocks. For the dsz aspect, we were able to successfully grow extra plates of Rhodococcus strain which was used to inoculate PCR tubes. The PCR did not go well, with significant streaking and false positives with similar banding pattern to previous gels run in the previous week. A final gel verification of a random sample of a tube of PCR products from dszA,B,C respectively and two negative control treatments involving master mix only and the lysed cells only illustrated the lack of discrepancy between the supposed successful amplification and the lysed cells (with lysate buffer) alone. Because of this we decided to take a different approach involving plasmid isolation carried out before PCR, rather than applying the PCR reagents directly to a lysed culture sample.
PSB1C3-hpaC verification through sequencing was successful, confirming the construction of our first biobrick. Subsequently, O/N cultures of the plasmid containing cultures were prepared and stored in glycerol at -80C. Furthermore, verification of catalase gene part (KatG-LAA, BBa_K137068)sent as a culture stab from the parts registry was initiated, with our newly identified biobricked-hpaC acting as a positive control, but the banding pattern was not very conclusive.
Week 8 (June 18 - June 22)
Decarboxylation
This week, we received the results of our gas chromatography of the eight tubes created last week, and unfortunately no hydrocarbons had been produced in any of them. It was thus very important for us to spend time researching why this was unsuccessful. We have found a number of things to consider. Firstly, the Washington iGEM team (creator of the petrobrick) has worked on system optimization, and has come up with an number of ways to increase alkane production. Information regarding this can be found here: https://2011.igem.org/Team:Washington/Alkanes/Future/Vector. There are many other considerations as well. We have received feedback on our sequencing results, and have been told that 87% is not high enough; we should be obtaining results of 100%. This is a problem because it presents the possibility that there is a mutation somewhere in our PetroBrick that is preventing effective operation. We may need to design inner primers to sequence the entire PetroBrick, as it is 2392bp and this may have prevented 100% sequencing results. We have also been using plastic falcon tubes, while hydrocarbon production has been tested in glass tubes. Incubation time and initial amount of cells to be innoculated are also concerns, as we have been having many problems with cell growth. The Washington Team often had an OD600 of 10 in their production media, whereas we have been tracking the OD of our initial broth only, so it is possible that the OD600 of our broth has been very low. As demonstrated by iGEM Washington, the OD600 is of utmost importance when it comes to hydrocarbon production, and is something critical that we need to improve. We are also considering extracting alkanes from our cells using HCl rather than ethyl acetate, as we many not be removing hydrocarbons effectively from the cells. We will consider each of these things when we attempt hydrocarbon production again.
Ring Cleavage
This week we started a new assay where Pf-5 was incubated with naphthalene. Since naphthalene is not volatile, GC headspace reading was not an appropriate method of measurement to indicate decrease of the compound. Instead we took OD600 readings to measure bacterial growth. The original cultures were taken from a Pf-5 culture in LB and inoculated into M9-MM without glucose. After 24 hours of growth these were subculutred into fresh M9-MM without glucose. This was to avoid carryover of glucose from the LB. Positive controls were also made that consisted of M9-MM with glucose and Pf-5. OD600 readings of the subcultures were taken over a period of 8 days. GC readings were done on the Pf-5 incubated with butylcyclohexane after 11 days and with toluene after 10 days. The trend remained the same as the measurements done after 5 and 6 days in the previous week. The amount of compound decreased in the experimental cultures but the amount of compound also decreased in the abiotic standards almost in equal proportions. With the results after 10 and 11 days we could see that the decrease was not due to the bacteria and we decided that the strain we were using did not contain the genes necessary for butylcyclohexane or toluene degradation. Throughout the week we continued the verification of XylE by performing a second colony PCR from the transformation plate using part J33204. The gel did not contain DNA of the right lengths and only the positive control worked.
Denitrification
The results of our transformation on CarAc and AntB from last week showed mostly red colonies indicating that the ligation was unsuccessful and that the PSB1C3 plasmid simply closed on itself with no insert. However, there were three colonies for CarAc that were white (cut with both X+P and E+P). Colony PCR was performed on these three colonies using biobrick primer sets to verify that the part had been inserted in these colonies, but unfortunately all PCR results were negative indicating an unsuccessful ligation.
We also performed a miniprep on Psudomonas putida strain #28 this week using a home-made protocol rather than the Qiagen kit. Miniprep A had a DNA concentration of 1539.8 ng/uL and Miniprep B had DNA concentration of 1001.2 ng/uL possibly indicating that there may have been a lot of genomic DNA contamination. However miniprep A product was still used as a PCR template for a reaction attempting to isolate CarAc, CarAd, AntA, AntB, and AntC. Special conditions for this PCR included replacing 60 uL of water with 60 uL of betaine heated to 37 degrees Celsius and running a Mg gradient ranging from 0.5 uL - 2.5 uL per tube for each gene. Of these only AntB had bands of the right size in 3 of the lanes (Mg concentrations of 1.5, 2, and 2.5 uL). However, all of the AntB bands (including the negative control) contained a contamination band at around 100 bases, probably due to the formation of primer dimers. This forced us to do a gel extraction rather than a PCR purification. This gel extraction did not work as the nanodrop results indicated high amounts of agarose contamination. Another PCR was performed using the same conditions, but using miniprep B instead as there was no more miniprep A product remaining. Unfortunately there were no bands for AntB, only the 100 basepair contamination bands.
Desulfurisation
PCR was reattempted on Rhodoccocus that was lysed using two different dilutions of the lysate buffer, but the gel verification confirmed the previous failure in using this approach. An alternative that involved preparation of an overnight culture of the Rhodococcus cells followed by a plasmid purification was followed. The plasmid purification eventually yielded plasmid samples with concentrations of 98.6ng/uL to 182.7ng/uL (4 samples obtained overall). Additionally, the catalase biobrick was used to transform some stock competent cells, and samples of some colonies were subsequently PCR'ed. Although, the gel verification showed some potential contamination, and the required banding patterns at around 2200bp was not obtained.
Week 9 (June 25 - June 29)
Decarboxylation
The week began initially with planning for continued attempts to produce hydrocarbons (Based on information that was learned using the University of Washington's "system optimization" techniques. We also backtracked to determine any additional sources of error within our procedures for the production media protocol. During the week, the sequencing results were examined in further detail, and it was determined that the entire gene sequence for aldehyde decarbonylase (ADC) was missing. This explained why our sequencing results for the petrobrick yielded only 87% complementarity. Based on this, there were two possible justifications for only having the acyl-acp reductase (AAR)gene and no aldehyde decarbonylase (ADC). Either the parts registry did not have the proper petrobrick (containing both AAR and ADC) in the assigned well in the 2012 kit plate sent to us, or we had extracted from the wrong well in the kit plate, assuming that it was the well containing the petrobrick. It was determined that the latter was true. As such, the transformation for the petrobrick had to be redone, and if gene sequencing yielded positive results (high complementarity), we would try the production media protocol once more in an attempt to get hydrocarbon production.
Desulfurization
PCR was attempted to amplify the genes of the dsz operon utilising an adapted PCR protocol with purified Taq polymerase that had been isolated from the host organism. Eventually, some banding pattern was obtained between 1200 and 1500 base pairs when a gradient thermocycler was used with melting temperatures ranging betweeen 55C to 65C. This was assumed to be indicative of successful amplification of dszB; however, further purification and gel verification results were inconclusive and no yield was obtained when placed tested using a nanodrop machine.
Ring Cleavage
After the 8 day incubation the readings of the naphthalene assay were graphed and there was a definite increase in the cell growth of the cultures containing naphthalene as a carbon source with indicated that the bacteria were most likely were metabolizing the compound. A verification of another part, Alkane hydroxylase system, from the registry was started. The system consists of 4 genes that have been shown to hydroxylate C5-C8 cycloalkanes. We thought that this part would assist in the degradation of cyclohexane and cyclopentane. The transformation was successful and the gel of the colony PCR showed one positive colony but since there was contamination in the gel we were not sure if we could conclude that the PCR had worked. Final GC readings on the butch and toluene cultures were taken and results were graphed.
Denitrification
This week we modified our PCR protocol to adjust for the homemade taq polymerase we were using. Template DNA was boiled for 10 minutes on its own to replace the initialization step. This is because this polymerase could not tolerate the 95 degree heat during the regular initialization step. Another modification we made was to boil our primers for 5 minutes prior to adding them to the master mix. This was done to discourage the formation of primer dimers. Finally we also attempted a touch down PCR which started at an annealing temperature greater than the Tm of our primers and gradually worked its way down to below the Tm of the primers. This was done to increase the specificity of our reaction and also eliminate the formation of primer dimers. Unfortunately these techniques still saw the formation of dimers, preventing us from performing a PCR purification. Again, the gel purification procedure did not lead to successful purification of our target gene. Due to the problems we have had so far our goal for next week is to perform an assay experiment to try and determine which of the Pseudomonas putida strains we have access to can actually degrade carbazole and therefore contain the pCAR1 plasmid.
Week 10 (July 2-July 6)
Decarboxylation
The main goal this week was to repeat the transformation and verification steps that were done previously with AAR for the petrobrick (submitted by the University of Washington). This involved transforming the petrobrick into E.coli, colony PCR for verification of a successful transformation, followed by mini-preparation and restriction digest. Results were submitted for gene sequencing. If the sequencing results come back as being successful (showing a high rate of complementarity), we will proceed forward with the production media protocol in an attempt to produce hydrocarbons.
Desulfurization
Top 10 E.coli cells were transformed with R0011 (IPTG inducible promoter in psb1C3 backbone), and resulting colonies were tested using cPCR. Colony PCR was performed on cells containing the catalase biobrick. Catalase is 2217bp long but since biobrick primers add about 200bp, bands of 2400 bp were expected if the part was present in the biobrick. These bands were observed, indicating that the KatG-LAA gene was most likely present.
PCR using Phusion high fidelity polymerase was carried out on dszA, dszB, and dszC in a gradient thermocycler. Amplification of non-specific bands was present for dszA and dszB, however strong banding for the desired size of the gene was observed for both (around 1500 for dszA, 1100 for dszB
Examining the sequences of the dszABC genes led to the discovery that all 4 had multiple illegal enzyme cut-sites in them that we have to eliminate before biobrick composite part construction can occur. dszA has four PstI cut sites, dszB has a PstI and a NotI and dszC has a PstI cut site. In order to eliminate cut sites, the Stratagene QuikChange mutagenesis procedure is going to be used, with the only alteration being that Kapa HiFi polymerase would be used during the process. Primers needed for the mutagenesis were designed based on the procedure mentioned above.
Ring Cleavage
This week we performed another colony PCR of 5 colonies from the transformation plate of part K398014 and ran the products on a gel. We obtained 4 positive colonies and made overnight cultures. We decided to stop using this part for the project and therefore did not require a further miniprep on these overnight cultures. However we made an overnight culture of the positive colony from the alkane hydroxylase system gel (colony 5) and miniprepped the overnight culture. This resulted in a good concentration of 345.6 ng/μL as we continued verification of XylE by doing two more colony PCRs of each part (K118021 and J33204) but only the positive control worked when we ran the gel.
Denitrification
The goal for this week was to grow different strains of Pseudomonas in media containing B-N media + glucose with only carbazole as a source of nitrogen. In theory only those strains which contained the pCAR1 plasmid would be able to use carbazole as a nitrogen source and thrive in this media. The strains we tested from glycerol stock included putida LD1, putida environmental strains 28-30, and Florescens PF5. Each of these strains had two cultures grown; one with carbazole and one without it. E. coli grown with carbazole was used as a negative control as it does not contain the pCAR plasmid. At timepoints of 22, 46, and 117 hours of growth samples were taken to do an OD600 reading to measure bacterial growth and had an ammonia assay performed on it. The detection of ammonia acted as a proxy for detecting carbazole degradation as it is an endproduct of the pathway. With this in mind an additional control strain of LD1 was grown in media containing glucose and ammonia. This is to detect the possibility that Pseudomonas can use ammonia as a source of nitrogen after it has degraded carbazole which would be problematic as far as using ammonia levels to determine carbazole degradation. So far we have been having difficulties reading the results of the ammonia assay with a plate reader and may resort to testing our samples using a different assay protocol next week. The OD600 readings were initially very high at 22 hours and then dropped at 46 hours.
Week 11 (July 9-July 13)
Decarboxylation
The sequencing results sent in for the petrobrick did not yield a high complementarity rate, therefore were unsuccessful. As such, we were not yet able to proceed with the production media protocol to attempt to produce hydrocarbons because we are not certain as to whether the petrobrick part has been isolated. It was then decided that the colony PCR would be redone with a new colony, followed again by a mini-preparation and restriction digest before gene sequencing occurs. If the sequencing results again yield a low complementarity rate, it can be deduced that the petrobrick sent to us in the 2012 kit plate does not contain a proper functional petrobrick.
Desulfurization
Following successful amplification of the dsz operon genes in the previous week, the genes were constructed into the PSB1C3 vector. Colony PCR verifications were observed to be positive. Furthermore, the insertion of part J13002 (pTetR)in front of the previously biobricked hpaC was attempted. Overnight cultures were also prepared using two colonies each for J13002 and R0011 (an IPTG inducible promoter that we hope to build in front of B0034). These cultures were then miniprepped to yield the respective parts.
PICTURE
Additionally, katG-LAA was built into a PSB1C3 backbone. The construction and availability of all these parts will be critical in the construction of our overall circuit for biodesulfurization. Colonies which looked good on cPCR were used to prepare overnight cultures, and were miniprepped and sent in for sequencing verification the following day. On the side, M9 minimal media was also prepared to carry out growth experimentation and overall desulfurization capability of Rhodococcus when exposed to DBT. The various growth treatments were M9 Media and glucose only, M9+glucose+DBT, M9+glucose+MgSO4+/-DBT, M9+glucose+MgCl2+/-DBT. 0.008g of FeCl2.4H2O was also added to each of the tubes. Samples were then inoculated with colonies of the Rhodococcus.
Denitrification
This week we took our last readings from the experiment that we started last week. The OD600 readings taken at 117 hours showed minimal growth over the weekend for all strains except for environmental strain #29 (with carbazole) which increased from 0.086 to 0.313. No other strain showed an increase of 0.100 or greater. This shows that only strain #29 was able to thrive in the carbazole media, possibly indicating that it contains the pCAR plasmid enabling it to use carbazole as a nitrogen source. To verify this we used a new ammonia assay protocol to measure the production of ammonia in the different cultures and theoretically how much carbazole was degraded. Unlike the previous assay we were able to produce a standard curve with a consistent slope based on samples with known amounts of ammonia. However, the only samples that showed any elevation of ammonia levels above the blank were the LD1 samples that were cultured with ammonia chloride to control for the possibility of pseudomonas using ammonia as a nitrogen source. The ammonia levels in this sample were relatively constant which would indicate that the LD1 was not using ammonia as a nitrogen source. Unfortunately the fact that none of the test samples had elevated ammonia levels did not give us any evidence that these strains contain the pCAR plasmid.
Ring Cleavage
Since we could not obtain any positive colonies from last weeks colony PCRs we began the transformation of each part again. The transformations were successful and when the colony PCR products were run on a gel there were two positive colonies for each part. Overnight cultures of the positive colonies were made. Minipreps were done on the overnight cultures and a restriction digest was performed on the products of the minipreps. The restriction digest products were run on a gel and there was one that looked like it had been successful but due to a poor quality ladder on this gel we could not make any final conclusions on the success of the restriction digest.
Week 12 (July 16 -July 20)
Decarboxylation
It was decided that the decarboxylation team would be split up into three individual components, a) to strictly work with the petrobrick, b) transformation and isolation of the aldehyde decarbonylate (ADC) and c) culturing the Nocardia strain NRRL 5646. With regards to the petrobrick, the sequencing results were once again unsuccessful and yielded a low complementarity rate. As such, it was decided that we would contact the University of Washington, and ask them to send us the petrobrick they composed and were working with in 2011. In addition, we also decided to produce our own biobrick identical to the petrobrick by using the acyl-acp reductase (which we had successfully transformed and isolated previously) with aldehyde decarbonylase (ADC). The ADC was transformed into the E.coli, and colony PCR was done in order to determine whether successful this was successful. This was followed by mini-preparation and restriction digest before gene sequencing occurred.
Ring Cleavage
We began the week by rerunning the restriction digest products on a gel with a good ladder and obtained one positive result with two bands. One band was around 1000 bp for the part and the other was at about 2000 bp for the plasmid backbone. We made a streak plate of the colony that had worked for the restriction digest and sent the miniprep product for sequencing. The colony that worked was from the transformation plate of part K118021. We also repeated a colony PCR of the transformants from part J33204 using 10 colonies and all of them looked like positive colonies. Overnight culutres were made and a miniprep and restriction digest was completed.
Denitrification
We received a new strain of Pseudomonas, LD2, this week that had previously been used to study the pCAR1 plasmid. It was grown up in PCA media and had its colonies used for colony PCR. We also received our new primers this week, further increasing the chances of successful PCR. Using the new strain and the new primers CarAa, CarAc, and CarAd all showed at least some bands of close to the correct size. After PCR purification we obtained DNA concentrations of 21.3 ng/uL for CarAa, 41.9 ng/uL for CarAc, and 32.8 ng/uL for CarAd. These products were then restriction digested using EcoRI + PstI sites.
Desulfurization
This week, while awaiting sequencing verification results which were required before we could begin the construction process, the desulfurization team initially aided in some of the tasks related to the other hydrocarbon groups. The success of the construction of BBa_J13002 with hpaC was also explored by using forward and reverse primers of R0040 (the promoter component of the composite part J13002). However, the eventual gel verification was inconclusive and sequencing results finally indicated an unsuccessful ligation. Additionally, the minimal media M9 preparation had been contaminated in the previous effort so this process was repeated to create tubes of each of the growth condition treatments detailed previously, and two repeats, one with an extra filtration step and one without was used to prepare the cultures.
Week 13 (July 23 - July 27)
Decarboxylation
The gene sequencing results for the aldehyde decarbonylase were unsuccessful. It was presumed that this occurred because the mini-preparation procedure being used may potentially be leading to a high contamination rate. As such, a colony PCR was done with a new colony for ADC, followed my mini-preparation using a different procedure, and then a restriction digest. The sequencing results now yielded a high complementarity rate and were considered successful. We then proceeded with making the biobrick consisting of the acyl-acp reductase (AAR) being ligated with aldehyde decarbonylase (ADC). Upon successfully forming this biobrick (ideally the same as a functional petrobrick), we will be able to test for hydrocarbon production using the general production media protocol. The University of Washington also responded, and said that they will be able to send us their functional petrobrick that was being used in 2011. With regards to culturing Nocardia, a “brain and heart infusion (BHI)” broth was used to inoculate the Nocardia in an attempt to get growth. Six pairs of mutagenic primers were also composed for carboxylic acid reductase (CAR), which is found in the Nocardia, as well as primers for the OleT enzyme.
Desulfurization
Mutagenic primers were redesigned after initial design was found to have premature stop codons. As part of the redesign process in constructing our overall gene circuits for desulfurization, a backbone switch of R0011 into a chlor-resistant vector was necessary, so this was attempted. The subsequent transformed products were plated on a Chlor plate and selected colonies were used to prepare O/N cultures, then miniprepped before finally being digested with enzymes EcoRI and PstIA. The resulting gel verification images were inconclusive as they did not show the required banding pattern around 50bp. Meanwhile, colony PCR was run on colonies transformed with katG-LAA constructed into a PSB1C3 backbone, as well as the hpaC+J13002 construct. katG was shown to have been successfully amplified, verifying its presence in the colonies, so overnight cultures were prepared and subsequently miniprepped. On the other hand, the construct was not successful so a third attempt was carried out. Colony PCR treatments that used either R0011 forward primers or B0034 primers were used and the overall constructs were made either on a chlor-resistant, or amp-resistant vector. Preliminary images of the gel verification appeared to have confirmed the construct, although sequencing verification will be the final indicator of overall success.
Denitrification
The restriction digested Car genes were ligated into vector PSB1C3 and transformed into competent E. coli cells. White colonies grew for CarAa and CarAc suggesting that ligation and transformation were successful, however no colonies grew for CarAd suggesting that the transformation did not work. Colony PCR was performed to confirm the presence of these genes within the vectors, however only CarAc was positive. The colonies containing the CarAc plasmid were then mini prepped and restriction digested to isolate the plasmid and send for sequencing. Further attempts at transforming CarAa and CarAd were not successful. Another goal was to perform colony PCR on the P. putida to isolate the AntABC genes using both the primers for the entire operon and those for the individual genes. No attempt showed any bands, although salt and temperature gradients were used as well as different types of polymerase. LD2 cultures were grown up to be plasmid prepped and genomic prepped in order to have a higher concentration of template DNA in the PCR master mix and more PCR will be attempted on these genes next week. Mutagenesis primers were also designed this week to remove the illegal restriction cut sites in the CarAd and AntB genes (NotI and 2xEcoRI respectively). Only 1 mutagenesis primer was designed for AntB as one of its EcoRI sites was very close to the beginning of the gene and could be removed using a new primer. During this process it was also discovered that the original Ant primers were designed for the wrong genes and therefore do not match the intended sequence. Although the new AntABC primers were designed using the correct gene the PCR may not be working due to the length of the operon so the design of new individual primers may be necessary. Finally, we also aimed to grow up the freeze dried culture of Rhodococcus erythroplois (source of the AmdA gene) that we received from DSMZ. The reactivation media was produced and autoclaved to begin the process next week.
Catechol Degradation
The colony PCR of part J33204 (XylE with a native rbs site) was done again and all five colonies looked like they contained the plasmid after running the products on a gel. After miniprepping and running a restriction digest on these five colonies two of them were sent to sequencing. An assay using the compound catechol was also started this week. E.coli cells (top 10) transformed with the part K118021 (XylE with a Pcst promoter) were used in this assay. Six overnight cultures were made, 3 using M9-MM and 3 using LB. These were spun down and resuspended in fresh M9-MM and LB and brought to a concentration of 0.1 M catechol by using a 1M stock solution. There seemed to be an initial colour change in the reactions from clear to yellow which was only visible in the cultures using M9-MM. All of the cultures were incubated overnight and they turned a black green colour. For this assay we should have used the supernatant because the reaction takes place outside of the cell so this is what we did next week.
Week 14 (July 30 - August 3)
Desulfurization
Sequencing results from the previous week's constructs were available confirming the contruction of KatGLAA in a chlor-resistant backbone, however, the backbone switch of R0011 also to PSB1C3 was not successful, probably owing to the small length of the RBS. On the other hand, the construction of J13002+hpaC was finally sent in for sequencing too, pending results. Site-directed mutagenesis of the dsz operon was also initiated - dszA has four PstI cut sites. dszB has a PstI and a NotI site. dszC has two PstI cut sites. Mutagenesis was started this week to change a single base pair in these genes in a way that the cut site is eliminated but the amino acid the gene codes for stays intact. In addition another mutagenesis has to be performed in order to replace the Tyr at position 63 of dszB gene with a Phe. Ohshiro 2007 showed that this mutation increases the activity of the enzyme. The mutagenesis was started by mutating the second PstI site in dszC (PstI2) and a control plasmid containing a point mutated β-galactosidase gene was also mutated with its specific primers. Kappa Hifi kit was used for all the mutagenesis. After the PCR mutagenesis and running a part of the PCR products on the gel, amplification was observed. Then the PCR products were DpnI digested to degrade the parental DNA and they were transformed. Control PCR products were plated on an amp plate containing IPTG and X-gal. The colonies that grew on the control plates were blue indicating that the mutagenesis had worked and the β-galactosidase gene was now functional. Then O/N culture of dszC mutants were miniprepped and digested with PstI enzyme and the results were successful (Lisa gel pic).
Attempts to do all the mutations in the genes in one step using the Knight procedure failed (http://openwetware.org/wiki/Knight:Site-directed_mutagenesis/Multi_site). This procedure used Taq ligase buffer. We suspected that the reason this procedure is not working might be that the Kappa polymerase is not functional in Taq ligase buffer. Therefore we did some experiments on the controls in Taq ligase kit and kappa polymerase kit to find out the buffer that they both work best in. The result was that both enzymes (kappa polymerase and Taq ligase) work best in a buffer made of half Taq ligase buffer and half kappa polymerase buffer.
Decarboxylation
<p>This week, we were pleased to find ample Nocardia iowensis growth on our BHI plate. We performed a PCR with our constructed CAR and NPT BioBrick primers to attempt to amplify these genes, so that we could insert them into BioBricks. This PCR utilized primers specific to our genes of interest. After running the gel, we found that we had ample amplification of both of these genes, as shown in the picture below: </html>In the above gel, lanes 4-5, 7, and 9-12 represent successfully amplifed NPT (669 bp). Lanes 14-16 and 18-20 show that CAR (4690 bp) has been successfully amplifed. Following this, we performed a Cycle-Pure Spin Protocol to purify the amplified CAR and NPT DNA. The resulting concentration of CAR was 171.6 ng/ul, while NPT was 20.8 ng/ul. Using a restriction digest, the CAR gene was cut with AscI and HindIII, while NPT was cut with both XbaI and PstI, as well as with EcorI and PstI. Using these same enzymes, the PET vector was cut to be ligated with CAR, and the PSB1C3 vector was cut to be ligated with both NPT digests. Following incubation, the inserts were ligated together with their vectors. Transformations were then performed, with the CAR/PET ligation transformed on a Kan plate, and the NPT/PSB1C3 E/P and X/P ligations transformed on chloramphenicol plates. Transformations were successful, but there were not a great deal of colonies on the CAR plate, and both NPT plates revealled a very high proportion of red colonies, indicating that our desired NPT gene had not been successfully ligated and transformed. Nonetheless, colony PCRs were run in order to test for successful transformations of the ligations the following week. Meanwhile, the results of the ADC gene sequencing came in, but unfortunately it was found that this gene did not correspond well to its theoretical sequence on PartsRegistry. It therefore could not be used with our AAR gene to produce a PetroBrick. The Washington iGEM team agreed to send us a plasmid stock of their PetroBrick and ADC.
Denitrification
We received the sequencing results this week for CarAc which came back as a 100% match to the expected nucleotide sequence. We also successfully ligated both CarAa and CarAd into PSB1C3 and transformed into top 10 E. coli cells. Colony PCR verified the presence of the biobrick plasmid in these colonies. These were both miniprepped, digested and sent for sequencing by the end of the week. Using a salt gradient, addition of DMSO, and the KAPA buffer specifically designed for GC-rich amplicons we were able to amplify the Ant operon from a genomic prep of LD2. However, all attempts at ligating and transforming this sequence were met with the unfortunate presence of red colonies. Next week we will recut a PSB1C3 vector with Xba and Pst to attempt to mitigate this problem. We also reactivated the R. erythropolis using DSMZ media #1 and grew it on plates made from DSMZ media #220 at 28 degrees. Colonies grew very quickly and were used for colony PCR and to make overnight cultures for downstream applications. The colony PCR using the amidase primers was unsuccessful, so a genomic prep was performed on the overnight culture. Unfortunately we lacked some of the required equipment and the genomic prep did not work, but with a few procedural modifications and the right equipment we are confident it will work next week. A glycerol stock of the erythropoplis was also made from another overnight culture. We also started construction on the CarAc part this week by inserting a ribosome binding site (B0034) in front of the gene. Finally, since the Ant operon was successfully amplified by PCR we designed mutagenesis primers for the 2nd EcoRI site in the operon rather than making new individual primers. We also designed a mutagenesis primer to remove the SpeI site in AmdA. Now that we have conclusively proven that the LD2 strain contains the pCAR1 plasmid we aim to demonstrate its ability to degrade carbazole and other nitrogen containing heterocycles using an assay experiment similar to the one performed in Week 10.
</html>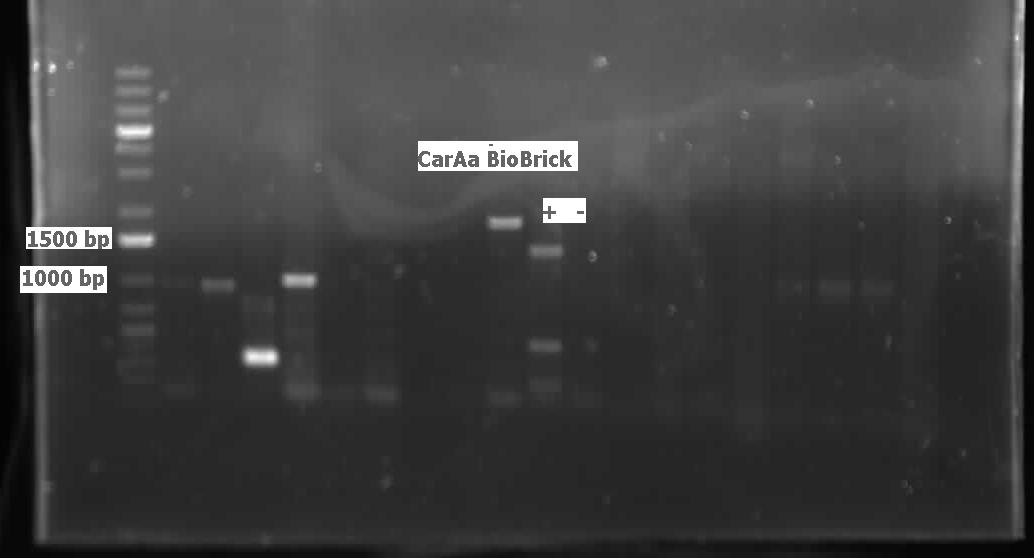
Catechol Degradation
The sequencing results for the part J33204 matched the part so a construction was started. The construction was a 3-way ligation putting the tetR promoter (R0040) and XylE (J33204) into a pSB1C3 vector. The ligation product was transformed into competent cells and a colony PCR was done on the transformants using both bio brick and R0040 primers. The primers for XylT, which is a ferredoxin that allows for XylE to be activated after a reaction with catechol, arrived and a colony PCR on four different strains of Pseudomonas putida was done. None of the strains seemed to contain the TOL plasmid, which is where XylT is found in certain strains of P. putida, because no bands showed up in the gel. The catechol assay was continued this week. Six overnight cultures were made, 3 using M9-MM and 3 using LB. They were spun down and the supernatant was brought to a catechol concentration of 0.1M, 0.2 M and 0.5 M using a 1M stock solution. The solutions turned light yellow initially, but after a few hours they turned brown. The assay was repeated with the same procedure as above but a control, a colony that did not contain the part K118021, was added. After the catechol was added to the supernatants there was a colour change from clear to yellow in all of the tubes, even the control. For a negative control catechol was added to M9-MM and this solution became pink. The tubes were incubated on the bench and checked regularly. The colour changed from yellow to pink to brown when they were left on the bench overnight.
Week 15 (August 7 - August 10)
Desulfurization
Sequencing results for J13002/hpaC returned negative, so a 3-way ligation method was used to retry the construction. The following parts were ligated with the restriction enzymes indicated in brackets after each: J13002(E/S) + HpaC (X/P) + PSB1K3 (E/P). Also, the more conventional construction of J13002(S/P) + HpaC(X/P) was reattempted. Furthermore, 3-way ligations were also attempted for B0034+KatGLAA+PSB1K3, and R0011+B0034+PSB1C3, as well as the two-way contruction of just KatGLAA after the B0034. After plating these transformations, colony PCRs were carried out on selected colonies and various samples that gave an indication on the gels of being successful were used to prepare O/N cultures then subsequently miniprepped.(insert gel images!) With regards to the site-directed mutagenesis side of the experimentation, dszA-PstI1 (the first PstI cut site in dszA) , dszB-PstI and dszC(PstI2 mutated)-PstI1 mutagenesis were performed following the procedure explained in the previous week. The gel below shows the successful result of digest confirmation. Multisite mutagenesis was repeated again using the modified buffer (half Taq ligase buffer and half Kappa buffer). However it was not successful again. We also tried doing multisite mutagenesis using pfu Turbo polymerase and following the Knight procedure without any modifications. No successful results were observed.
Decarboxylation
We ran the gel on the CAR and NPT colony PCRs from last week, but unfortunately these gels were blank. Another colony PCR was performed, with 12 colonies from each of the following plates: CAR/PET, NPT(X/P), NPT(E/P). Both NPT plates did not look very promising, as they were completely covered by red colonies, with only a few white ones. Many of the white colonies from the previous week had become red. 8 overnight cultures were also performed so that a restriction digest could be done on the CAR colonies, to see if the part was there. These overnight cultures were prepared with the Kan antibiotic. Another ligation was done with the three combinations of cut inserts and vectors: CAR with PET, NPT (E/P) with PSB1C3, and NPT (X/P) with PSB1C3. Restriction digests were performed on the CAR overnight colonies with AscI and HindIII, and when the gel was run, a single lane was shown to contain both the CAR gene and PET vector, as shown below:
</html>As shown in the gel above, lane 3 indicates 2 bands, the uppermost at about 5.4kb, identifying as PET. This single lane also contains a clear band of CAR, a bit under the 5000bp mark, at 4690bp. The ladder used was 1 kb plus. This successful result allowed us to proceed with mutagenesizing the 6 illegal cut sites of CAR, including: 2 PstI sites, 1 EcorI site, and 3 NotI sites, with the primers that we had designed. Two additional colony PCRs of NPT were performed, but unfortunately a band indicating NPT was not visible in any of the traisl, even when an additional plate of both the NPT (E/P) and NPT (X/P) restriction digests were ligated and transformed. Very few white colonies could be seen on any of the transformations, but we were determined to continue trying. We then attempted to mutagenesize the first illegal cut site of CAR, which would be a PstI site. The mutagenesis primers needed to be diluted and stock solutions were made. Then, we attempted to mutagenesize the PstI site of CAR with that site's specific primers, using various concentrations of template DNA (5ng, 20ng, 50ng). We used the KAPA system to perform this plasmid amplification, using the thermocycler settings of the Desulfurization team. The mutagenesis PCR would be run over the weekend to hopefully have successful results.
Denitrification (or are we calling it denitrogenation now??)
This week we received the sequencing results for CarAa and CarAd. CarAd was a match, however CarAa did not match at all and had misplaced biobrick restriction enzyme sites. Due to this we decided to restart the biobricking process for CarAa all the way back to PCR from the plasmid. By the end of the week we had progressed to the point of transforming it into competent E. coli cells, however a thick lawn of bacteria had grown overnight making it impossible to pick colonies. A streak plate was made from this plate to attempt to get less dense growth. Since the sequencing came back positive for CarAd the next step was to perform site directed mutagenesis on its NotI restriction enzyme site using the primers that were designed previously. The PCR was successful, showing very bright bands at 3000 bp, and the PCR product was digested with DpnI to remove parental DNA. Transformation of this product will indicate whether the mutagenesis was successful. We were also able to successfully perform a genomic prep of Rhodococcus erythropolis and amplify the AmdA gene this week. It was ligated into PSB1C3 and transformed into competent E. coli by Friday, and a confirmation colony PCR was run on Friday evening to verify that the gene was inserted. These results will be available by Monday. After many attempts the Ant operon was finally ligated and transformed as well this week. Confirmation PCR showed bands of about 3000 bp which is consistent with the size of the operon. By Friday the successful colonies had been grown up and miniprepped, ready to send for sequencing early next week. The CarAc/B0034 construct was also successfully transformed this week and verified by running a PCR using the B0034 forward primer. It was also miniprepped and is ready to send for sequencing next week. In addition to this work constructing biobricks we also started an experiment to demonstrate the function of the pCAR plasmid in LD2. Strains of LD2 were grown up in LB and then washed and placed in B-N media + glucose and various experimental additions. These additions included carbazole, 4-PBAH (a nitrogen containing naphthenic acid), ammonia chloride, and a negative control with no nitrogen source. Strains of E. coli were also grown under the same conditions to compare growth and ammonia production of LD2 to a strain that does not contain the pCAR plasmid. These results should act as a preview of what we expect our final biobrick containing E. coli species to produce as well as allowing us to establish LD2 as a positive control for this experiment.
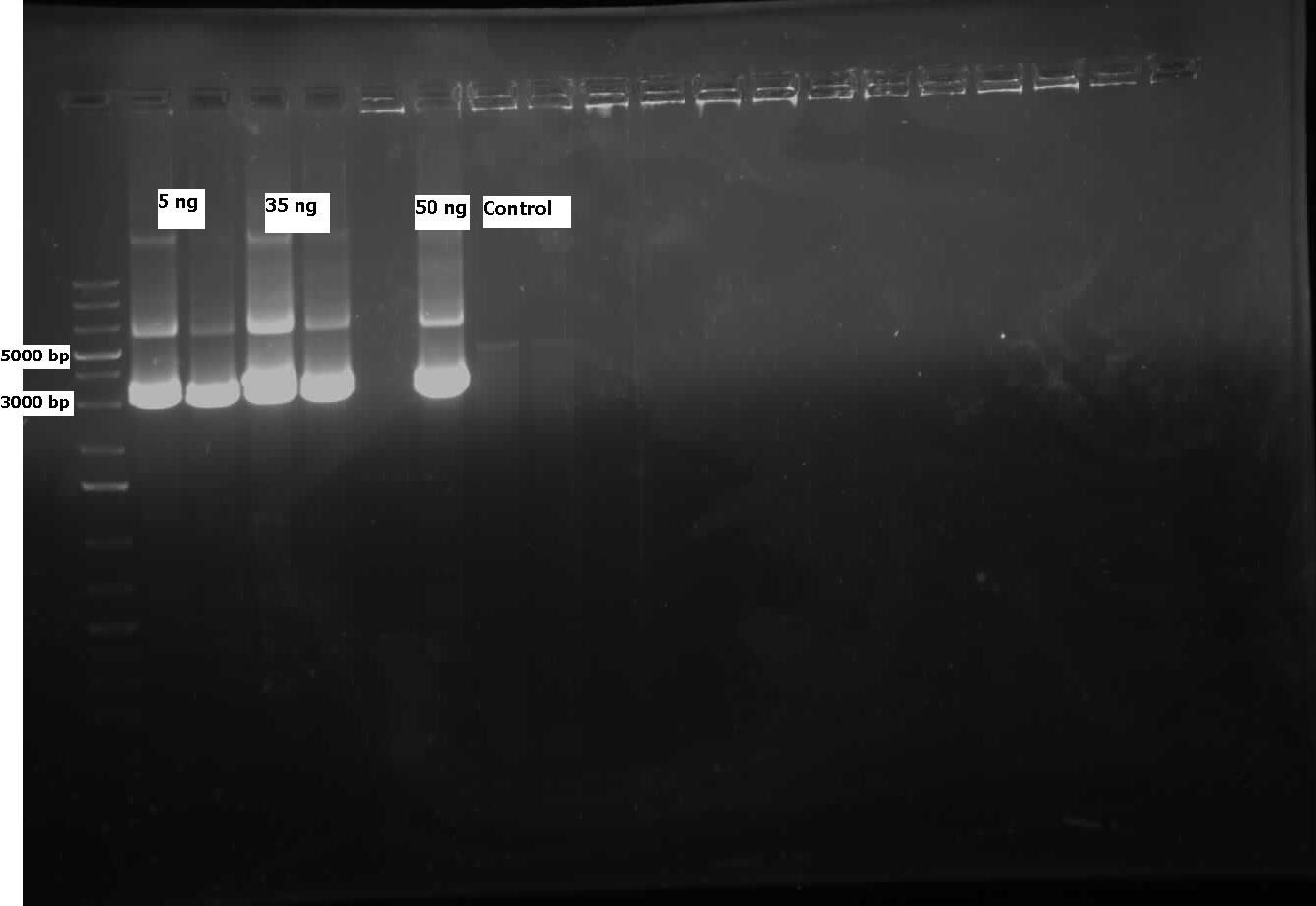
Week 16 (August 13 - August 17)
Desulfurization
Following on from the progress in the previous week of the mutagenesis, dszB(PstI mutated)-Y63F and dszA(PstI1 mutated)-PstI3 mutagenesis were performed. The gel below shows the digest confirmation.
Another approach was tried to do mutagenesis with a quicker pace. In this approach, after the PCR mutagenesis, the PCR products were purified. Afterwards they were incubated with T4 PNK and ligase. After heat inactivating ligase and T4 PNK, the products were DpnI digested. Subsequently another round of DNA purification was performed. However, the results were unsatisfactory after the digest confirmation. Sequencing results came back. dszA (PstI1 and PstI3 mutated) and dszB(PstI and Y63F mutated) were good. However dszC (PstI1 and PstI2 mutated) had an insertion next to the PstI1 cut site. Mutagenesis was repeated on the dszC(PstI2 mutated). dszB(PstI and Y63F mutated)-NotI and dszA(PstI1 and PstI3 mutated)-PstI4 mutagenesis were performed. The gel below shows the digest confirmation. To investigate the actual desulfurisation capability of our initial Rhodococcus source culture for the dsz operon, a desulfurization assay was prepared by innoculating various treatments of the M9 media prepared previously. Also, a conditioning agent composed of 100ml of 95% ethanol, 50ml glycerol, 30ml of 12M HCl (aq) and 70g of NaCl(s) was prepared. The assay relies on the turbidity of a sample containing sulphate ions which are precipitated (hence the turbidometric nature of the assay) upon adding BaCl2(s). Elsewhere, B0034 was constructed with dszC and of the colonies that grew on the plate of the transformed products, colony PCRs were performed and the gel image gave an indication of it being successful. However, further analysis through sequencing showed that the constructions were not successful, and so the constructions were restarted.</p>
Denitrogenation
The sequencing for the CarAc/B0034 construct came back with only CarAc and no evidence of a B0034 site preceding it. The confirmation PCR that was run last week using the B0034 forward primer seemed to indicate that it was inserted, however since B0034 is only 12 base pairs the primer may not have been specific enough. The construction on this part was restarted this week. CarAa colony PCR on last week's transformation showed two positive colonies, however upon culturing, miniprepping, and digesting these colonies the plasmid appeared to be too large (3000 base pairs as opposed to the expected 2000). This may be due to a problem of a mislabeled plasmid tube that has been experienced by other team members, causing the gene to be inserted into PSB1AC3 rather than PSB1C3. To test this, a streak plate was made from these colonies and grown on an ampicillin resistant plate. Mutagenized CarAd was successfully transformed into top 10 E. coli cells, cultured, and miniprepped. The miniprep product was digested with NotI to test whether the NotI site was successfully removed. These results were good, as the plasmid ran at 2000 base pairs and the insert at just under 1000 base pairs in the mutagenized CarAd whereas the non-mutagenized CarAd had its insert cut into 2 bands. This will be sent for sequencing next week to confirm the mutation. Unfortunately the sequencing results came back negative for the Ant operon meaning we will need to restart genomic PCR on it next week. Multiple transformations and confirmation PCRs were run on AmdA this week, however none displayed a band size of 1700-1800 base pairs as expected. We have continued to try different digested plasmids as other team members also had difficulties this week with ligation into PSB1C3, possibly indicating a problem with our antarctic phosphatase stock.
The other news from this week was the final results from our carbazole degradation experiment attempting to demonstrate LD2's ability to degrade carbazole and hopefully other nitrogen containing compounds. Unfortunately, these results were very difficult to make sense of as the LD2 that were cultured with carbazole grew, but did not show elevated ammonia levels which would be indicative of carbazole degradation. One of our controls was LD2 cultured with ammonium chloride to test whether LD2 was capable of using ammonia as a nitrogen source. This would pose a problem for us as we were planning to use an increase in ammonia levels as a proxy for carbazole degradation rather than measuring carbazole degradation directly using GC-MS. However, the LD2 showed significantly less ammonia after 44 hours when compared to 26 hours indicating that it may be degraded, either by the LD2 or by an unaccounted for abiotic process. This has lead us to decide to use GC-MS and directly measure carbazole levels in our sample the next time we do this experiment.
Decarboxylation
This week, the attempted PstI mutagenesis of CAR was run on a gel. Unfortunately, amplification was unsuccessful, but we had an idea for why this might have occurred. It was discovered that our plasmid of 9kb would need a 7.5 minute extention time, rather than the 1 minute extention time that we had originally programmed into the thermocycler. The mutagenesis experiment was repeated, this time with a 7.5 minute extention time. When these results were analyzed with gel electrophoresis, we had bands appearing to indicate success with the 50ng samples of CAR that we had run, as shown below.
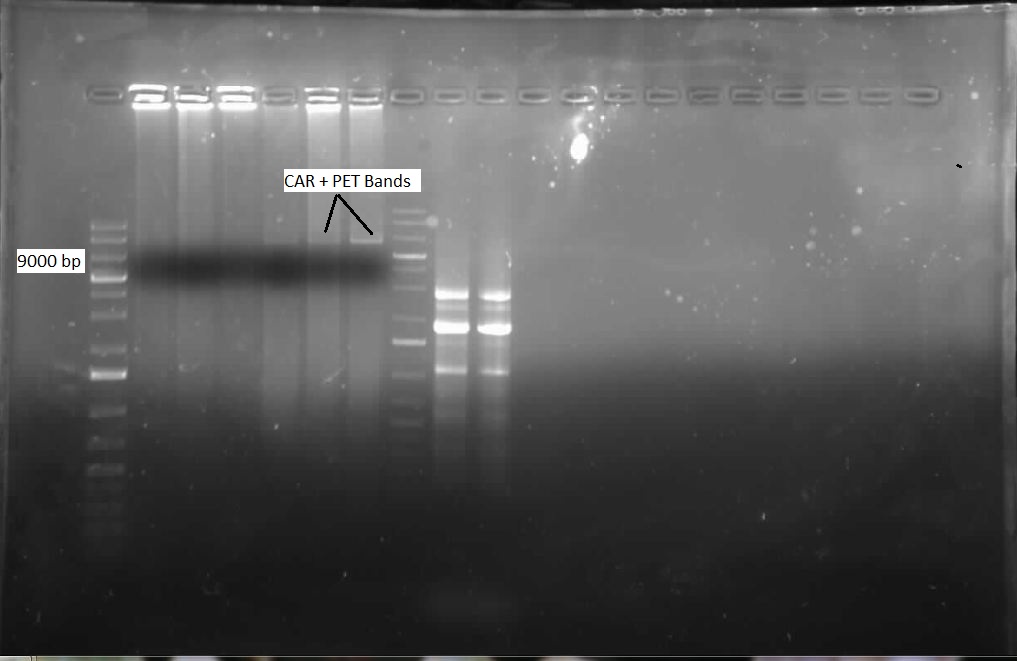
After combining these two PCR tubes and incubating with DpnI, we performed a transformation with this DNA and allowed the plate to incubate overnight. Also, we continued to try and verify NPT, but were once again unsuccessful. When we viewed the plate that we had transformed the following day, it was found that we did not have any colonies. We presumed this was because the 9kb plasmid was very difficult to uptake with its large size. Several more transformations were attempted. We also attempted a new procedure called site-directed mutagenesis, which would use a single mutagenesis primer per site, rather than one for each strand, and supposedly had the capacity to mutagenize multiple illegal cut sites at once. We would try to mutate 3 sites with this protocol: the 2 PstI sites, and 1 EcorI site. The PCR was run overnight, with 8 different samples, at two different concentrations of DNA (50ng and 80ng), with four different annealing temperatures. The following day, each sample was incubated with DpnI for 6 hours, and then each was tranformed on a kan plate. A restriction digest of the NPT plasmid was once again performed, cutting with both (E/P) and (X/P) as before, this time using more DNA. An overnight culture of a single potentially successful NPT colony was made, to perform a restrictin digest to see if NPT was present - this would be done over the weekend. Finally, a transformation of the single mutagenesis KAPA PCR of PstI was performed, to hopefully yield more colonies.
Week 17 (August 20 - August 24)
Decarboxylation
This week, we carried forth with the second and third mutagenesis sites of the CAR gene. Our new plasmid with our first PstI site was PCR amplified with the Kapa system and our EcorI mutagenesis primers to carry out the second mutation, to remove the EcorI illegal cut site from our gene. After the PCR was complete, we ran our product on a gel to see if the amplification worked. Bands indicated success, as shown in the gel below. Following this verification, we mixed our two seemingly successful PCR tubes with 1 uL of DpnI, and left them to incubate for one hour. The PCR product was then transformed into highly competent E.coli cells, and the plates were left overnight in the 37 degree incubator. Unfortunately a mistake prevented colonies from growing, but the process was repeated and our second attempt yielded many colonies. We made overnight cultures of 8 of these colonies, with 4 from each plate, with Kan as the antibiotic for selective growth. The next day, a miniprep was performed on each overnight culture to isolate the CAR+PET plasmids that had hopefully undergone their second mutation, of their single EcorI site. Once the plasmid isolation was complete, we performed a restriction digest using EcorI to cut, in order to verify that the illegal cut site had been removed and the plasmid could no longer be digested with this enzyme. Our success is demonstrated in the gel below. We immediately proceeded with the third mutagenesis, which would this time be performed on the second (and last) PstI site. Using our new plasmid now with two illegal cut sites removed, we performed an amplification PCR, using our PstI(2) primers and the Kapa system. After running this on a gel, we determined bands of success, as shown below. We then once again treated our PCR amplified tubes with DpnI for one hour, and then transformed into highly competent E.coli. The plates were left overnight to allow colony growth. Our second accomplishment of the week was successfully amplifying the OleT gene from Jeotgalicoccus, by using the Kapa system. This was very successful, as shown in the gel below. We then performed the cycle-pure spin protocol to remove contamination from our PCR product, and then proceeded to perform a restriction digest on the product with HindIII and AscI so that we could ligate it into the PET vector. After this ligation was complete, it was transformed with E.coli on a Kan plate, and left overnight to incubate. Unfortunately we do not see colonies yet, but the incubation is not complete yet, and might still be successful. Thirdly, we obtained the PetroBrick stock from the Washington team on Tuesday, and immediately proceeded to transform it. We obtained a great deal of colonies, and then performed colony PCR on 12 of these colonies to ensure that our ~2400bp PetroBrick was present in these cells. Last but not least, we have still been working on a successful purification of NPT from a transformed plate. A colony PCR indicated that NPT was potentially in a few of these colonies, so we made overnight cultures of these four colonies, and performed a miniprep the next morning, and then a restriction digest to verify that the gene was present.
Denitrogenation
The CarAa was re-streaked onto ampicillin plates and grew, probably showing that it was ligated into PSB1AC3, not PSB1C3, which explained the irregularity of our confirmation digest last week. It was sent for sequencing, but in case it was incorrect we also started a new CarAa pipeline by PCRing it from the Pseudomonas genome again and redigesting and ligating. It was transformed into PSB1C3 over the weekend. CarAc did not see a lot of progress this week as we spent much of the time attempting to construct a RBS (B0034) in front of it, but were unsuccessful. CarAd was also subjected to this construction with B0034 and we were able to send it for sequencing by the end of the week. Multiple new constructs were also started for CarAd in case the sequencing is negative. The Ant operon was successfully transformed and confirmed by colony PCR this week, after which it was grown up and sent for sequencing. We also began to attempt mutagenesis on one of the EcoRI sites in the operon, but had no amplification of the plasmid. We also re-digested some PCR purified product to start another ligation in case sequencing failed. After many attempts we were also able to finally successfully transform an AmdA/PSB1C3 plasmid into top 10 cells this week. After a miniprep they showed bands of about 1800 base pairs on the confirmation digest gel, so they were also sent for sequencing this week and had mutagenesis started on the illegal SpeI site. Unfortunately, the sequencing sent early in the week came back with ambiguous results. The forward reaction for AntABC was a match, but its reverse reaction matched to CarAa. Both CarAa reactions matched nothing. Since these two genes are over 20 000 base pairs apart on the template plasmid it is virtually impossible that they were actually combined into one plasmid (PCR extension time would not have been long enough and even if it was the product would have run much above the 3000 base pair mark). This is likely a mistake on Eurofins end, so we are not 100% sure if we have 1 good gene, both of them, or neither. Also the sequence for mutagenized CarAd showed only about the first 100 base pairs, which were correct however not long enough meaning it was a poor reaction and inconclusive. Finally, we also re-started our carbazole degradation assay using the GC-MS at biological sciences. This process will take 2 weeks to demonstrate LD2's ability to degrade carbazole. We are forcing it to use carbazole as a carbon and a nitrogen source in one sample and as only a nitrogen source in another sample (this sample has had glucose added to it).
Desulfurization
This week, progress was made in determining the desulfurization activity of our Rhodococcus strain as measured by the sulfate release using a turbidometric assay. WE encountered numerous challenges in our prescribed protocol as the concentrations that we are using to prepare out standard curve may be too dilute, or the composition of out conditioning agent may be flawed. Additionally, steps were taken to determine the decomposition of DBT to 2-HBP through GCMS analysis, but due to a preparation error, the DBT was added to a growth solution of M9 media prematurely and the autoclaving process decomposed the DBT releasing a yellow colouration into the solution. These two approaches in determining the desulfurization capability of the dsz operon will be further investigated.
Since the dszC second mutagenesis had proven to be unsuccessful last week, the dszC(PstI2 mutated)PstI1 mutagenesis was repeated. Also dszA(PstI1,3,4 mutated) PstI2 mutagenesis was performed. dszA and dszC were sent for sequencing on Wednesday. dszB was sent for sequencing on Friday. Sequencing results of dszA and dszC were back by Friday. dszC was successful. However, dszA contained an insertion next to the binding site of PstI4 cut sit, so the last two mutations must be redone. dszB(PstI and Y63F mutated)-NotI-mutagenesis was also repeated in case the result of the sequencing was not successful. These constructions were repeated. J13002-dszB and B0034-dszC constructions were attempted, however they were not successful as indicated by cPCR. Constructions of J13002/hpaC were carried out and also came back negative in sequencing, however B0034/katG-LAA was sequence confirmed.
Week 18 (August 27 - August 31)
Decarboxylation
This week, the primary objectives from last week were continued. Another restriction digest of OleT and then a ligation was performed, in order to see if we could obtain more colonies following transformation. After preparing overnight cultures and minipreps of eight of these colonies, we attempted to cut the OleT/PET vector with EcorI to verify that the gene had successfully inserted into the vector. Unfortunately, we did not have any luck, even after several attempts. Our attempts to verify this will continue. We also performed ligations with NPT (cut with XbaI and PstI) with JI3002 (cut with SpeI and PstI), so that we could produce a biobrick with JI3002 before NPT to act as a promoter and ribosome binding site. We decided on how we will build our two primary constructions, and where each part will be. Also, we attempted to amplify the CAR gene in the PET vector (with 3 mut sites replaced) with biobrick primers, so that we could get it inserted into a biobrick plasmid. After several tries, this amplification was successful.
Desulfurization
dszB sequencing results came back as successful. dszA(PstI1,3 mutated)-PstI2-mutagenesis was performed and sent for sequencing. Also dszA(PstI1,2,3 mutated)-PstI4-mutagenesis was performed, and this was also sent for sequencing.
Constructions of J04500 with hpaC, dszB, and katG-LAA were performed. Transformations were carried out at the end of the week.
Denitrogenation
The new stock of CarAa that we had PCRed from LD2 was sent for sequencing this week and came back as a match, so we have begun constructing a RBS site and a TetR promoter (J13002) in front of it. At this point all the Car genes have been successfully amplified, inserted into a biobrick plasmid, and sequence verified. We also continued to attempt to construct CarAc with an RBS site (B0034) in front of it, but had no success. On a better note, we confirmed this week that CarAd was mutated properly and we were also able to insert an RBS site in front of it. The next step for this gene will be to attempt a plasmid switch as it is currently in PSB1A3 and needs to be in PSB1C3 for submission to the parts registry. After sending a new batch of the Ant operon for sequencing we got the same confusing results as we did last week. We therefore hypothesized that our primers were annealing to a homologous region that is similar to the Ant genes and very close to CarAa. This means that we need to restart PCR on this gene as all of our current product is incorrect. The sequencing results for AmdA that came back this week were also somewhat confusing as they did not quite match the sequence we expected. However, they were a 100% match for the AmdA gene in a different strain of Rhodococcus erythropolis. This means that DSMZ probably sent us the wrong strain of bacteria and that we were very lucky that the primers we designed based off of the expected template actually worked to amplify a slightly different gene in another strain. The good news is that this new sequence contains no illegal cut sites, which explains why mutagenesis was not working (there was nothing to mutate) and allows us to immediately begin construction of J13002 in front of AmdA this week.
Week 19 (September 3- September 7)
Denitrogenation
CarAd/B0034 was successfully plasmid switched into PSB1C3, making it ready for submission. Construction of J13002 and CarAa may have been successful as the digest of the miniprep looked promising. It will be sent for sequencing next week. Unfortunately there was no progress made on the constructions of AmdA with J13002 or CarAc with B0034. These will be redigested and religated for next week. We could not amplify the Ant operon despite trying many variations of PCR using high fidelity KAPA polymerase. This is probably due to the multiple, large regions of homology on the operon that the reverse primer binds to making amplification difficult. Because the function of this gene can theoretically be replaced by AmdA, we may scale back efforts dedicated to it, as the chances of successful amplification appear slim.
Week 20 (September 10 - September 14)
Denitrogenation
Constructs that continued this week included AmdA with J13002, CarAa with J13002, and CarAc with B0034. Some sequencing was sent, but no positive results were obtained. A new approach to biobrick the Ant operon was implemented this week. A new forward primer was designed that would anneal about 1500 base pairs upstream of the Ant operon, avoiding the region of homology that is repeated at various points on the plasmid. Once this region is successfully amplified we could use this amplified DNA as a template for another PCR reaction which would use the normal Ant primers. Since the template would no longer include regions of homology these primers should be free to work properly and amplify the Ant operon. The PCR using the new primers was started this week, and although amplification at 4500 base pairs was observed as expected, there was also a lot of non-specific amplification. This means that to purify the amplified DNA that we want and avoid the other products we will have to perform a dreaded gel extraction next week.
 "
"