Team:Calgary/Notebook/Killswitch
From 2012.igem.org
Kevinm.huie (Talk | contribs) |
Kevinm.huie (Talk | contribs) |
||
| (61 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | {{Team:Calgary/TemplateNotebookOrange| | ||
| + | TITLE=Team Killswitch| | ||
| + | CONTENT = | ||
<html> | <html> | ||
<!-- | <!-- | ||
| Line 5: | Line 8: | ||
Patrick. | Patrick. | ||
--> | --> | ||
| - | |||
<h2>Week 1 (May 1-4)</h2> | <h2>Week 1 (May 1-4)</h2> | ||
| Line 15: | Line 17: | ||
<p>The mechanism of death is still settled upon the micrococcal nuclease, but regulation of the death genes will be difficult. Existing repressible promoters in the registry still tend to be leaky in their expression, but we need to test this out. We may look into the TetR-repressible promoter (<a href="http://partsregistry.org/Part:BBa_R0040">R0040</a>) and also the clλ regulated promoter (<a href="http://partsregistry.org/Part:BBa_R0051">R0051</a>).</p> | <p>The mechanism of death is still settled upon the micrococcal nuclease, but regulation of the death genes will be difficult. Existing repressible promoters in the registry still tend to be leaky in their expression, but we need to test this out. We may look into the TetR-repressible promoter (<a href="http://partsregistry.org/Part:BBa_R0040">R0040</a>) and also the clλ regulated promoter (<a href="http://partsregistry.org/Part:BBa_R0051">R0051</a>).</p> | ||
| - | <p>We also found a few riboswitches responsible to various metal ions such as Mg<sup>2+</sup> and Mn<sup>2+</sup>. The <i>mgtA</i> riboswitch activates translation of the death gene when magnesium ions are not present in the solution. The <i>mntA</i> riboswitch deactivates translation of the death gene when manganese ions are not present. Considering this, we may be able to find a method of precipitating or otherwise sequestering Mn<sup>2+</sup> ions out of the tailings water prior to entering the bioreactor. This way, our bacteria would not die when they come into contact with the manganese in the tailings water. Possible additives include carbonate (CO<sub>3</sub><sup>2-</sup>) or hydroxide (OH<sup>-</sup>), though this may alter the pH | + | <p>We also found a few riboswitches responsible to various metal ions such as Mg<sup>2+</sup> and Mn<sup>2+</sup>. The <i>mgtA</i> riboswitch activates translation of the death gene when magnesium ions are not present in the solution. The <i>mntA</i> riboswitch deactivates translation of the death gene when manganese ions are not present. Considering this, we may be able to find a method of precipitating or otherwise sequestering Mn<sup>2+</sup> ions out of the tailings water prior to entering the bioreactor. This way, our bacteria would not die when they come into contact with the manganese in the tailings water. Possible additives include carbonate (CO<sub>3</sub><sup>2-</sup>) or hydroxide (OH<sup>-</sup>), though this may alter the pH greatly.</p> |
| - | <p>One other proposal would be to use promoters that activate when the bacteria detect the formation of a biofilm | + | <p>One other proposal would be to use promoters that activate when the bacteria detect the formation of a biofilm, for example, on glass beads. Engineering a monolayer of cells on a bead means that if a cell detaches, its death genes will activate. Further research into this is needed.</p> |
<h2>Week 3 (May 14-18)</h2> | <h2>Week 3 (May 14-18)</h2> | ||
| Line 25: | Line 27: | ||
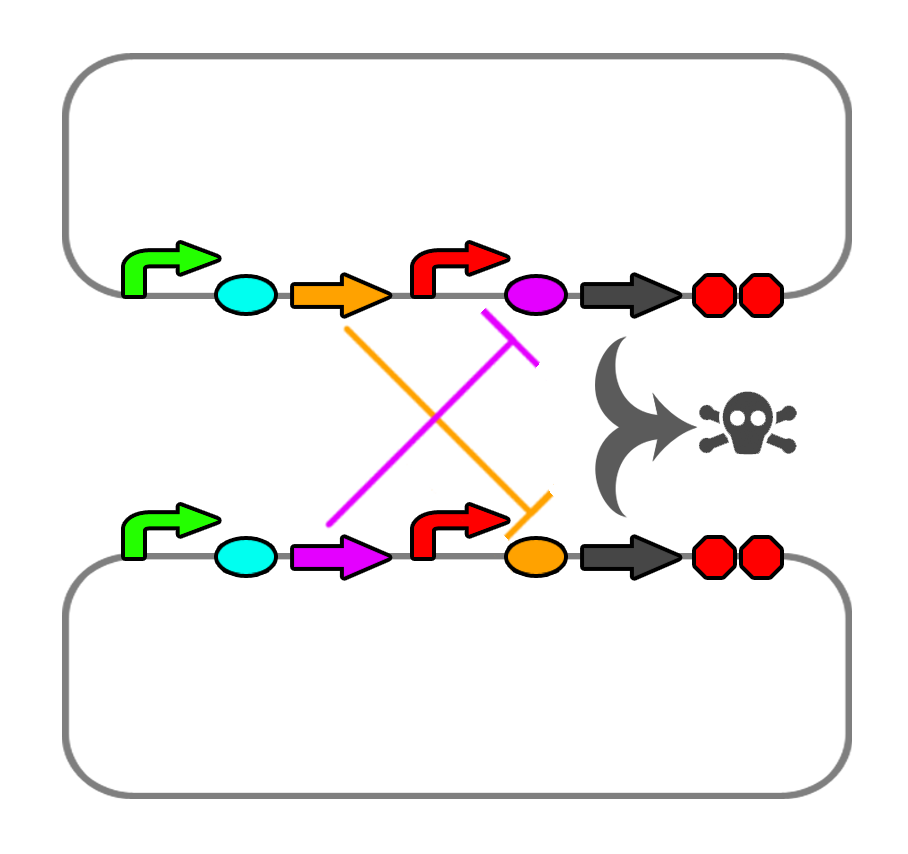
</html> [[File:Ucalgary_2012_Codependence_Mechanism_V2.png|thumb|400px|center|Proposed method of codependence. Two plasmids in two separate strains will produce ligands that repress the riboswitch controlling each other's death genes.]]<html> | </html> [[File:Ucalgary_2012_Codependence_Mechanism_V2.png|thumb|400px|center|Proposed method of codependence. Two plasmids in two separate strains will produce ligands that repress the riboswitch controlling each other's death genes.]]<html> | ||
| - | <p> We also found a glucose repressible promoter that we could have potentially used upstream of the kill switch. This promoter is found in <i>F. tularensis</i>. However we also found that this promoter is not found in <i>E. coli</i> and <i>F. tularensis</i> has a unique polymerase which used this promoter. Hence using this promoter does not seem feasible. | + | <p> We also found a glucose repressible promoter that we could have potentially used upstream of the kill switch. This promoter is found in <i>F. tularensis</i>. However we also found that this promoter is not found in <i>E. coli</i>, and <i>F. tularensis</i> has a unique polymerase which used this promoter. Hence using this promoter does not seem feasible. |
Next week we will be looking into further possibilities for the kill switch. | Next week we will be looking into further possibilities for the kill switch. | ||
| Line 31: | Line 33: | ||
<h2>Week 4 (May 21-25)</h2> | <h2>Week 4 (May 21-25)</h2> | ||
| - | <p>In terms of our literature search we decided against the glucose repressible promoter from F. tularensis because it was not native to E.coli and upon contact with the authors of the paper we found that the promoter did not work in E.coli. We also came upon two systems that we are interested in further investigating. The first one was a glucose activated promoter that was found in the registry which we | + | <p>In terms of our literature search we decided against the glucose repressible promoter from F. tularensis because it was not native to E.coli and upon contact with the authors of the paper, we found that the promoter did not work in E.coli. We also came upon two systems that we are interested in further investigating. The first one was a glucose activated promoter that was found in the registry which we are thinking of coupling to an inverter and the second one is a rhamnose-based regulatory system.</p> |
<p>On Tuesday, in addition to our literature research, we decided on some of the circuits that we will be building over the next couple of weeks and came up with a plan detailing what will be done each day. These are the circuits: | <p>On Tuesday, in addition to our literature research, we decided on some of the circuits that we will be building over the next couple of weeks and came up with a plan detailing what will be done each day. These are the circuits: | ||
| Line 39: | Line 41: | ||
<p> On Wednesday we proceeded with the transformation of 3 parts from the iGEM kit into our E.coli: a tetR promoter, a GFP-double terminator, and a RBS-GFP-double terminator. | <p> On Wednesday we proceeded with the transformation of 3 parts from the iGEM kit into our E.coli: a tetR promoter, a GFP-double terminator, and a RBS-GFP-double terminator. | ||
| - | On Thursday we verified our transformation process with a cPCR where we ran 10 colonies from each plate with our controls. We ran a gel | + | On Thursday we verified our transformation process with a cPCR where we ran 10 colonies from each plate with our controls. We ran a gel of the PCR products and viewed it to verify the transformation and to decide which of the successful colonies to plate and grow overnight. </p> |
</html> [[File:UCalgary_Wrong_Gel_1.jpg|thumb|400px|center]]<html> | </html> [[File:UCalgary_Wrong_Gel_1.jpg|thumb|400px|center]]<html> | ||
| Line 54: | Line 56: | ||
<p> | <p> | ||
| - | During the week of | + | During the week of May 28th, the killswitch team made progress toward transforming/verifying nine different parts in <i>E. coli</i>. Work began with continuation of the previous week's transformation of R0040 (TetR promoter), I13401 (GFP terminator), and I13504 (RBS GFP terminator). Note that these three parts will be necessary for characterization of the four different kill system triggers being explored by the killswitch team. Verification procedures (ie, colony PCR) showed successful transformation of I13401. Results for the other two parts were inconclusive and cloning of R0040 and I13504 will continue next week. |
</p> | </p> | ||
| - | |||
| - | |||
| - | |||
<p> | <p> | ||
| Line 97: | Line 96: | ||
<p>Hence new primers were designed. </p> | <p>Hence new primers were designed. </p> | ||
| - | <p> | + | <p> PhoPNEW-AS CTGCAGCGGCCGCTACTAGTAGCAGTAATTTTTTCATCAGCGCAATTCG</p> |
| + | <p> PhoQNEW-AS CTGCAGCGGCCGCTACTAGTAGCATATTTATTCATCTTTCGG </p> | ||
| + | <p> mntRNEW-S gaattcgcggccgcttctagGAGGAAGCACAATGAGTC </p> | ||
| + | <p> mntRNEW-AS CTGCAGCGGCCGCTACTAGTAGCGTGCGTAAAAAAGG</p> | ||
| - | <p> | + | <p>The killswitch team also focused on verification of both the magnesium/manganese (MgtA promoter, MgtA riboswitch and MntP promoter) and registry parts (R0040, I13401 and I13504).</p> |
| - | <p>For the magnesium/manganese parts this week we insert them in biobrick vectors in order to carry out colony PRR. Colony PCR of the magnesium/manganese parts could not have been done first since we did not have the right primer, therefore by doing a construction digest we could use the standard biobrick primers. From the gel, the results were inconclusive as the ladder was not clear and the parts were too small to tell the difference between them and primer dimers, therefore next week goal is to perform another CPCR of the parts.</p> | + | <p>For the magnesium/manganese parts this week we insert them in biobrick vectors in order to carry out colony PRR. Colony PCR of the magnesium/manganese parts could not have been done first since we did not have the right primer , therefore by doing a construction digest we could use the standard biobrick primers. From the gel, the results were inconclusive as the ladder was not clear and the parts were too small to tell the difference between them and primer dimers, therefore next week goal is to perform another CPCR of the parts. As well as doing colony PCR of the MgtA promoter, a restriction digest of this part was done. The gel showed good results as the bands were bright and the right size.</p> |
| + | |||
| + | </html>[[File:U.calgary.2012_06.08.2012_MgtA_Promoter_Restriction_Digest_2.jpg|thumb|400px|center|Restriction Digest of MgtA Promoter]]<html> | ||
<p>For the registry parts R0040 no results were shown in the initial colony PCR, however another gel was done with R0040 from the 2010 and 2011 registry plate. The colony PCR of R0040 from 2011 showed the rights bands, however the results are inconclusive as the negative was contaminated, therefore most likely another colony PCR needs to be done.</p> | <p>For the registry parts R0040 no results were shown in the initial colony PCR, however another gel was done with R0040 from the 2010 and 2011 registry plate. The colony PCR of R0040 from 2011 showed the rights bands, however the results are inconclusive as the negative was contaminated, therefore most likely another colony PCR needs to be done.</p> | ||
| - | <p>The registry part I13401 had a successful week as both the colony PCR and the | + | <p>The registry part I13401 had a successful week as both the colony PCR and the restriction digest verified our parts. Since the result was successful we performed glycerol stocks of the plasmid and prepared it for sequencing the following week.</p> |
| - | </html>[[File:U.calgary_2012.06.04.2012-CPCR_Registry_Parts.jpg|thumb|400px|center|Colony PCR of I13401|]]<html> | + | </html>[[File:U.calgary_2012.06.04.2012-CPCR_Registry_Parts.jpg|thumb|400px|center|Colony PCR of Registry Part I13401]]<html> |
| + | |||
| + | </html>[[File:U.Calgary.2012_06.05.2012-Emily_gelb_Registriction_Digest_I13401.jpg|thumb|400px|center|Restriction Digest of Registry Part I13401]]<html> | ||
<p>The part I13504 showed no results this week therefore the goal next week is to find another part that is similar to I13504 and test its verification.</p> | <p>The part I13504 showed no results this week therefore the goal next week is to find another part that is similar to I13504 and test its verification.</p> | ||
| + | |||
| + | <h2>Week 7 (June 11-14)</h2> | ||
| + | |||
| + | <p> | ||
| + | |||
| + | This week we continued the insertion of magnesium promoter (MgtA) into biobrick plasmid (PSB1C3) to carry out the colony PCR to confirm the presence of the Mgta part in the plasmid using Bbk_CP_F and Bbk_CP_R primers. We are currently in the process of further verification by using a mini prep. | ||
| + | We also transformed four registry parts (J06702 [RBS-mCherry-dTerm], I13507 [RBS-RFP-dTerm], E0840 [RBS-GFP-dTerm] and E0240 [RBS-GFP-dTerm]) directly from the kit plates and verified the parts using colony PCR the following day. The primers used to verify these parts were Bbk_CP_F and Bbk_CP_R. These parts were successfully transformed but we still saw some contaminant bands in the gels (see below). | ||
| + | </p> | ||
| + | <p> | ||
| + | |||
| + | </html>[[File:CPCRmntPpromoterUCALGARY.jpeg|thumb|400px|center]]<html>]</p> | ||
| + | |||
| + | <p> | ||
| + | |||
| + | Transformation of MntP in PSB1C3 vector was verified using colony PCR. However, the results indicated that the presence of MntP as well as a contaminant band around 3000 bp.</p> | ||
| + | |||
| + | <p></html>[[File:FourGlowingReportersUCLAGARY.jpeg|thumb|400px|center|Colony PCR of Registry Parts where J04650 was successful]]<html>]</p> | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | Transformation of registry parts I13521 [TetR-B0034-RFP-dTerm], I13507 and J04650 [RFP-dTerm] were performed and colony PCR was carried out to verify the presence of the parts in the cells. J04650 was the only part that was successfully transformed. Though the other parts were not successfully transformed, we can still see non-specific bands in the gel (see below). We are currently doing mini prep to additionally verify the success of transformation.</p> | ||
| + | |||
| + | </html>[[File:06.13.2012-transformedpartsHGD.jpg|thumb|400px|center|Colony PCR of Registry Parts where J04650 was successful]]<html> | ||
| + | |||
| + | <h2>Week 8 (June 18-June 22)</h2> | ||
| + | |||
| + | <p> This week we did a restriction digest of all the plasmids that were isolated. We confirmed that we had J04650, I13507, E0240 and mgtA riboswitch. Hence these were sent to sequencing. We got sequencing results back and confirmed that we have J04650, I13507, J06702, R0040, E0240 and mgtA riboswitch. Henceforth, we started constructing the mgtA riboswitch with J04650. </p> | ||
| + | |||
| + | </html>[[File:06.21.12 Mgta Construct Digest Himika(1).jpg|thumb|400px|center|Restriction digest of mgtA riboswitch and J04650 construction]]<html> | ||
| + | <p> The mgtA riboswitch with J04650 was confirmed using PCR as well as restriction digest. We plan on sequencing it when we are able to construct it with a promoter. </p> | ||
| + | |||
| + | |||
| + | <h2>Week 9 (June 25-June 29)</h2> | ||
| + | |||
| + | <p> This week we attempted to construct the mgtA riboswith and J04650 construct with promoters such as MgtA and TetR. We also tried to plasmid switch I13507 such that we can put mgtA promoter on to that composite part. So far we have verified through colony PCR that it worked. This was verified again with restriction digest however the gel did not look too good therefore we ran the inserts and vectors to see how they looked. We will reattempt this again next week. </p> | ||
| + | |||
| + | <h2>Week 10 (July 3-July 6)</h2> | ||
| + | |||
| + | <p> This week, we attempted to religate our constructions from the previous week as they were not verified as positive in our colony PCR. We had run a gel to see if the inserts and vectors were digested properly and they were. Following transformation, we verified it through cPCR. The two gels show the results of construction involving two promoters (Mgta Promoter and a constitutive tetR promoter, R0040) and the mgta rbsw with different reporter gene constructs from the previous week. There are also some plasmid switches and constructions of the promoters and just the riboswitch. Based on these results, we isolated plasmids of R0040 + (Mgta RBSW and I13401) from L6 and L7 as well as I13507 from L8 from the first gel and R0040 + (Mgta RBSW and J04650) from the second gel (circled in red). We also transformed B0034, a ribosome binding site and constructed Mgta rbsw with a reporter construct with fast degrading GFP. </p> | ||
| + | |||
| + | </html>[[File:iGEMCalgary2012.07.04.2012 Construction cPCR.jpg|thumb|400px|center|Gel 1: Colony PCR of Constructions]]<html> | ||
| + | </html>[[File:iGEMCalgary2012.07.04.2012 Construction cPCR 2 (1) with label.jpg|thumb|400px|center|Gel 2: Colony PCR of Constructions]]<html> | ||
| + | |||
| + | <p> We verified these constructions with a digest. The third gel shows the digest and the lanes circled are the ones that look good and were sent to sequencing. </p> | ||
| + | |||
| + | </html>[[File:iGEMCalgary2012.07.06.2012 verification of overnight digest with labels.jpg|thumb|400px|center|Verification digest of constructions]]<html> | ||
| + | |||
| + | <h2>Week 11 (July 9-July 13)</h2> | ||
| + | |||
| + | <p> During the week of July 9th, construction was done on R0040 with J04650 and K082003 constructs as well as E0240 and B0034. Similarly the MgtA promoter was constructed with either J04650 construct, K082003 construct, E0240 or B0034. Furthermore a gel was run on the J04650 and K082003 constructs to check for genomic contamination which showed positive results for sequencing. </p> | ||
| + | |||
| + | </html>[[File:Uofcalgary_07.11.2012_MINI_PREP_RESULTS_2.jpg|thumb|400px|center|Gel of the Mini Prep to check for genomic contamination]]<html> | ||
| + | |||
| + | <p> From the restriction digest of the construction R0040 or MgtA promoter with the J04650 or K082003 constructs little results was shown, however some of the sequencing presented varying results. Sequencing of the R0040 with MgtA riboswitch and J04650 worked beside for weird sequencing around the scar site while both registry part E0240 and B0034 were good. The construct with MgtA promoter did not work as sequencing showed that the promoter was not part of the construct therefore the construction of these parts will have to continue another week. </p> | ||
| + | |||
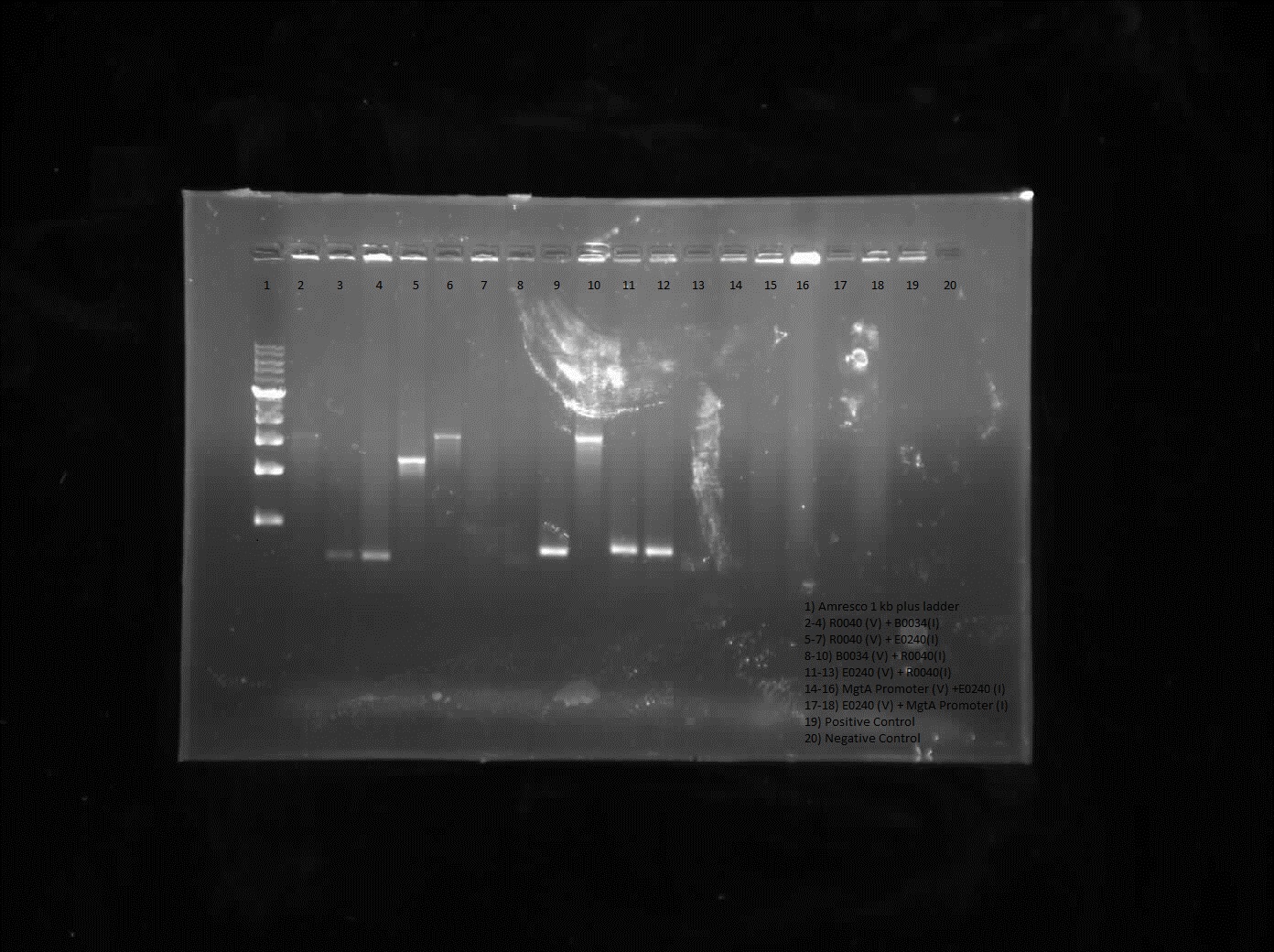
| + | <p> Results from colony PCR showed positive bands for R0040 (vector) with B0034 (insert), R0040 (vector) with E0240 (insert) and B0034 (vector) with R0040 (insert). </p> | ||
| + | |||
| + | </html>[[File:U.Calgary.2012.07.12.2012_kevin_%26_rai_%27s_july10_constructs_CPCR_2.jpg|thumb|400px|center|Colony PCR of R0040 constructions]]<html> | ||
| + | |||
| + | <p> Construction of these constructs will be done another week as the restriction digest did not look good at the same time the ladder was not readable. </p> | ||
| + | |||
| + | </html>[[File:U.Calgary.2012.07.13.2012_CPCR_overnight_digest_and_RD_2.jpg|thumb|400px|center|Unreadable gel due to the ladder smearing]]<html> | ||
| + | |||
| + | |||
| + | |||
| + | <h2>Week 12 (July 16 to July 20)</h2> | ||
| + | |||
| + | <p>During week twelve (July 16th to July 20th), the kill switch team worked on three different projects: first, transformation/verification of kill genes; second, transformation and PCR of parts for the rhamnose inducible kill system; and third, construction of parts for characterization of the magnesium riboswitch kill system.</p> | ||
| + | |||
| + | <p>The commercially synthesized kill genes K131009 (colicin E2 operon w/o SOS promoter) and S7 nuclease (micrococal nuclease gene) were transformed into E. coli. Colonies transformed with these genes, along with the previously transformed K117000 (for colicin activation), K112808 (holin-antiholin lysis system), and endolysin, were subjected to verification procedures including colony PCR and verification digests. Successful transformants were sent off for commercial sequencing.</p> | ||
| + | |||
| + | <p>The team also began work on the the rhamnose inducible kill system since primers for rhaR and rhaS genes were received along with the synthesized rhamnose promoter. Temperature gradient PCRs (54C-59C; also 54C-65C with phusion)for the rhaR and rhaS genes were performed with no success. The rhamnose promoter was transformed into E. coli of which a plasmid stock was isolated by miniprep.</p> | ||
| + | |||
| + | <p>Finally, the team continued with constructions necessary for characterizing the magnesium riboswitch kill system. They continued with verification of the MgtA riboswithch with GFP and also GFP(+LVA tag) under a constitutive promoter. | ||
| + | </p> | ||
| + | |||
| + | |||
| + | |||
| + | <h2>Week 13 (July 23 to July 27)</h2> | ||
| + | |||
| + | <p>During the week thirteen, the killswitch team performed plasmid switches of S7, mntP riboswitch and pRha (rhamnose inducible promoter) from the commercial IDT synthesis vector into pSB1C3.</p> | ||
| + | |||
| + | <p>Additionally, the team attempted to remove RhaR and RhaS from the E. coli genome via PCR. Kappa polymerase was used with a 50-65 degrees Celsius temperature gradient with no success. Given this failure, touchdown PCR from 58 to 74 degrees Celsius was performed on the RhaR and RhaS genes. These strategies failed. The team re-analyzed the primers used in these protocols; the antisense primers were erroneous and had to be redesigned.</p> | ||
| + | |||
| + | <p>Additionally, we tried construction of mgtAp with mgtArbsw-K082003. However no positive clones appeared in the follow up verification.</p> | ||
| + | |||
| + | <p>We also worked on construction of R0040+B0034, R0040+E0240, R0040+MgtA Riboswitch, R0011+MgtA Riboswitch, MgtA Promoter+B0034, MgtA Promoter with MgtA Riboswich and GFP(+LVA tag), and B0034 with GFP(+LVA tag). These constructions are all in different phases of the verification process. The team also attempted to expedite construction of pRha (rhamnose promoter) and S7 (micrococcal nuclease) with BOO34 by building the directly from the commercial IDT synthesis vectors; however, this process failed due similar resistance genes on the IDT and BOO34 vectors.</p> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <h2>Week 14 (July 30 to August 3)</h2> | ||
| + | |||
| + | <p>This week we tried to construct R0040 and R0011 with mgtArb-K082003. This construction was done both ways, i.e both R0040 and mgtArb-K082003 were used as both vectors and insert. Out of these 2 constructions, R0040-mgtArb-K082003 gave positive clones however R0011-mgtArb-K082003 did not. A sample from a single colony with R0040-mgtArb-K082003 in pSB1C3 was sent to sequencing; the results were postively verified.</p> | ||
| + | <p>Additional constructions that were started include the following:</p> | ||
| + | <ul> | ||
| + | <li><p> R0011-mgtArbs</p></li> | ||
| + | <li><p> R0011-mgtArbs-K082003</p></li> | ||
| + | <li><p> mgtArb-S7</p></li> | ||
| + | <li><p> pRHA-B0034</p></li> | ||
| + | <li><p> B0034-S7</p></li> | ||
| + | <li><p> R0011-B0034</p></li> | ||
| + | <li><p> mntPrbs-K082003</p></li> | ||
| + | <li><p> mntPrbs-S7</p></li> | ||
| + | <li><p> B0034-S7 (ie, S7 in pSB1C3)</p></li> | ||
| + | </ul> | ||
| + | <p>Digest verification showed success with K082003+B0034, J13002+K082003, R0040+ MgtA Riboswitch, and R0040+E0240; henceforth these were submitted for sequencing.</p> | ||
| + | |||
| + | |||
| + | |||
| + | <h2>Week 15 (August 6 to August 10)</h2> | ||
| + | |||
| + | <p>Colony PCR was performed on constructions from the previous week. Positive results were obtained with the following constructions: R0011-MgtAriboswitch, MgtAriboswitch-S7, MntpRiboswithch-S7, R0011-B0034, and MntpRiboswich-K082003. Minipreps and verification digests were performed from the relevant colonies.</p> | ||
| + | |||
| + | <p>The commercially synthesized Mntp riboswitch and moco riboswitch were switched into PSB1C3 vectors. Colony PCR and restriction digest verification yielded positive results; these samples are destined for sequencing verification. Additionally, the team constructed tetR-RBS-GFP(+LVA. Verification procedures were performed. TetR-RBS-GFP(+LVA) is awaiting sequence verification.</p> | ||
| + | |||
| + | <p>Temperature gradient PCR and salt gradient PCR were used to optimize and amplify the moa operon from the E. coli genome. The rhaR gene was amplified from Top Ten E. coli colonies and and biobricked in pSB1C3 with Kapa polymerase. The rhaS gene was amplified from Top Ten E. coli on a 52 to 67 degree Celsius temperature with Kapa polymerase and DMSO.</p> | ||
| + | <p> | ||
| + | Continued working with MgtA Promoter+ B0034, R0011 +MgtA Riboswitch,B0034+K082003 and trying to get DNA of the MntP Promoter.</p> | ||
| + | |||
| + | <h2>Week 16 (August 13 to August 17)</h2> | ||
| + | |||
| + | <p>For the week of August 13th continuation of the construction of the MgtA Promoter +B0034, R0011+MgtA riboswitch and B0034+K082003 was conducted. Constructions of MgtA Promoter+B0034 and B0034+K082003 were tested with plasmid PCR several times however none of the plasmid PCR showed any bands on the gel. R0011+MgtA riboswitch restriction digest did not work therefore a reconstruction was done and will follow thought for the next week. One of the colonies from MntP Promoter IDT plate digested well and was therefore plasmid switch into pSB1C3.</p> | ||
| + | <p> We started plasmid switching parts that were not in pSB1C3 into pSB1C3. Additionally, we also started to characterize the magnesium system using mgtAp-mgtArb-K082003, mgtAp-mgtArb, R0040-mgtArb-K082003. We also tried to construct R0011-mgtArb, mntPrb-S7, mntPrb-S7J13002-K082003, R0011-mgtArb-K082003, B0034-S7. At the end of the week we were able to sequence verify B0034-S7 and mntPrb-K082003. </p> | ||
| + | |||
| + | <p>Finally, the rhamnose promoter system has been reoriented:</p> | ||
| + | </html>[[File:PrhaFinal.png|thumb|700px|center]]<html> | ||
| + | |||
| + | <p>In this finalized version of the construct, the RhaR gene has been left out from the system. Given that RhaR activates RhaS, and RhaS activates the pRha promoter, it should theoretically be sufficient to constituitively express the RhaS promoter to activate the system. Construct must continue so that it can be tested whether this new design will function and be repressible with glucose. Check out the <a href="https://2012.igem.org/Team:Calgary/Project/HumanPractices/Killswitch/Regulation#rhamnose">project page</a> for more details. | ||
| + | </p> | ||
| + | |||
| + | |||
| + | <h2>Week 17 (August 20 to August 24)</h2> | ||
| + | |||
| + | <p>For the week of August 20th most of the constructions were redone while we continue with any other constructions we have already started since we noticed that we haven’t fully heat kill the enzymes. MgtA Promoter + B0034 was constructed with another B0034 that was in a kan vector while the MntP Promoter was redone from IDT as the previous MntP Promoter was not working.</p> | ||
| + | <p> This week we tried to construct mgtAp-mgtArb-S7-mgtArb so that we can construct CviAII when it would arrive. </p> | ||
| + | |||
| + | <p> We also attempted redoing the nuclease assay with S7 and <i>E. coli</i> genomic prep.This time the three different amounts of S7 was used. We used 200U, 50U and 10U. </p> | ||
| + | |||
| + | <p> The moa operon was biobricked and sequence verified. We are trying to construct the moa operon with a costitutive promoter and a ribosome binding site. </p> | ||
| + | |||
| + | </html>[[File:S7nuclease troubleshooting.png|thumb|500px|center|There is degredation with S7 and it looks like 10U is optimal for our purposes]]<html> | ||
| + | |||
| + | <p>Constructions of the rhamnose promoter (pRha) with GFP and S7 continued. These constructions have proven challenging and will have to be repeated next week.</p> | ||
| + | |||
| + | |||
| + | <h2>Week 18 (August 27 to August 31)</h2> | ||
| + | |||
| + | <p>For the week of August 27th constructions of MgtA Promoter +B0034, MntP Promoter +B0034 in Kan, R0010+ MgtA Riboswitch, R0010 +MntP Rbsw + S7, MntP promoter + MntP Riboswitch + S7 had to be redone as the ones done on last Friday did not grow or verification through PCR did not show positive results. After repeat trails of these construction CPCR showed that positive bands for MntP Promoter plasmid switch into kan vector, MgtA Promoter+B0034, MntP Promoter +B0034 (Kan), R0010 +MgtA Riboswitch and R0010 + MntP Riboswitch + S7. Since there was positive results was shown for these constructions, mini prep and digestion verification will be done the following week.</p> | ||
| + | |||
| + | <p> This week we kept reattempting our constructions however sequencing came back as things it should not have been randomly. Mostly Car genes from the denitrogenation team. Therefore we had to back to our glycerol stocks from the previously verified part and try to reconstruct our parts.</p> | ||
| + | |||
| + | <h2>Week 19 (September 3 to September 7)</h2> | ||
| + | |||
| + | <p>For the week of September 3rd constructions (MntP Promoter plasmid switch into kan vector, MgtA Promoter+B0034, MntP Promoter +B0034 (Kan), R0010 +MgtA Riboswitch and R0010 + MntP Riboswithc + S7), was verified with restriction digest and sent to sequencing. Also MntP Promoter+ MntP Riboswitch +S7 construction was reattempted after the enzyme incident where they were not fully heat killed.</p> | ||
| + | |||
| + | |||
| + | <h2>Week 20 (September 10 to September 14)</h2> | ||
| + | |||
| + | <p> The moco riboswitch was biobricked and sequence verified. Constructions of the moco riboswitch with different promoters (R0010, R0011, R0040) were attempted along with the moco riboswitch with the GFP LVA tag, CviaII and S7. The attempts of construction of the moa operon with the constitutive promoter and a ribosome binding site have not been successful thus far and is an ongoing project.</p> | ||
| + | |||
| + | <p> Some of the constructions of pRha and GFP looked promising in colony PCRs and verification digests. These samples were sent for sequencing. In the meantime, we attempted to induce the potential colonies with rhamnose to see if there was evidence of a successful construction prior to sequencing results. Results were negative compared to constituitively expressed GFP and a negative control. We awaiting sequencing results to confirm this failure.</p> | ||
| + | |||
| + | |||
| + | <h2>Week 21 (September 17 to September 21)</h2> | ||
| + | |||
| + | <p> The constructions with the moco riboswitch and the moa operon were not successful and were reattempted this week. </p> | ||
| + | |||
| + | <p> Rhamnose promoter constructions with GFP were negative as shown in sequencing results. We recommenced with this construction.</p> | ||
| + | |||
| + | <h2>Week 22 (September 24 to September 29)</h2> | ||
| + | |||
| + | <p> The constructions with the moco riboswitch and the kill genes and the GFP with LVA tag looked promising on the gels and were sent to sequencing. We tried to construct those constructs with a promoter. R0010 with the moco riboswitch and CviaII looked promising on the gels and was also sent to sequencing. </p> | ||
| + | |||
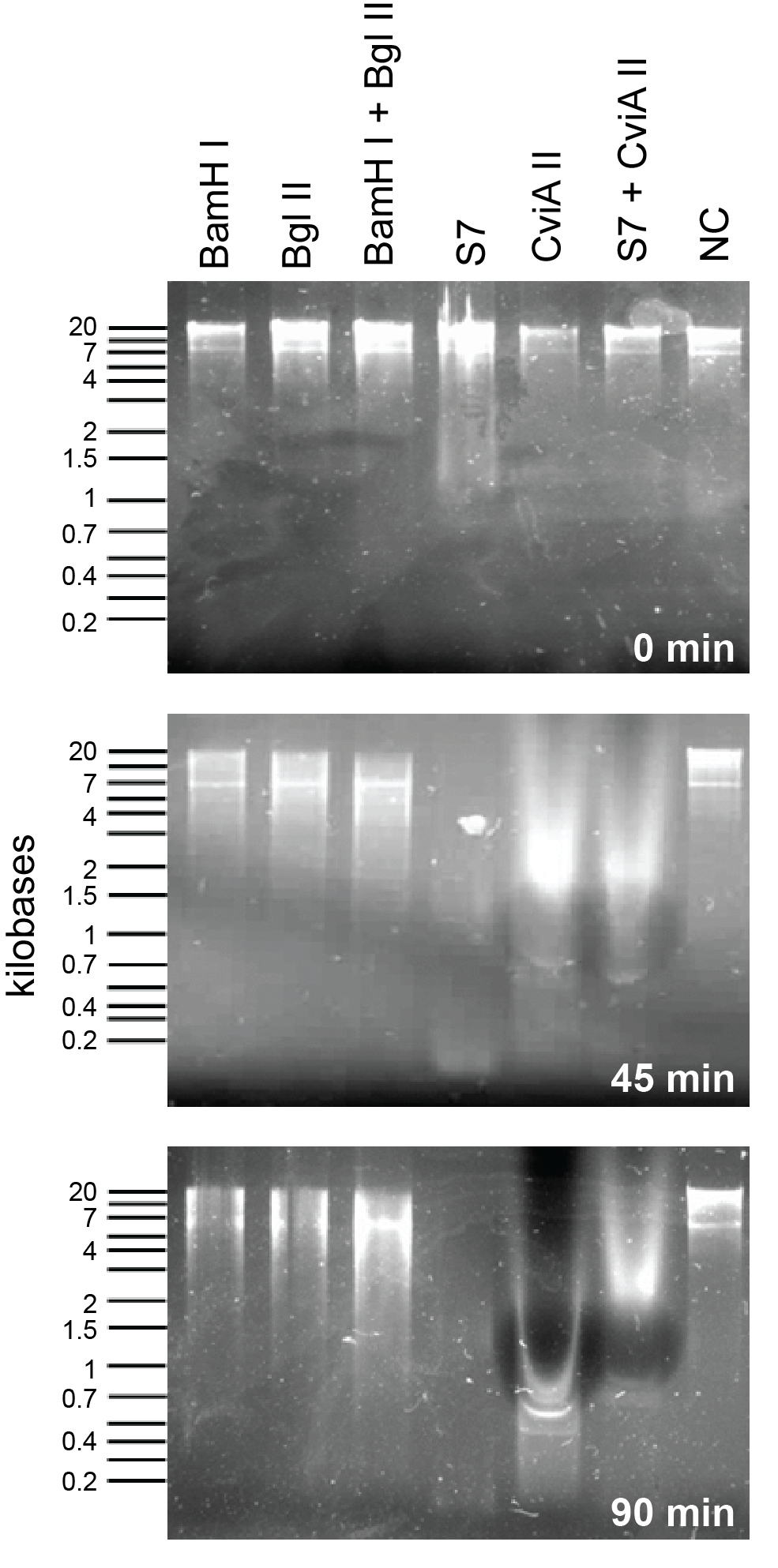
| + | <p> This week we reattempted the nuclease assay to compare BglII, BamHI, S7 and CviAII against each other. This time we used 10U of enzyme in each condition and tested degradation at every 45 minutes. </html>[[File:UCalgary2012 RE-S7&CviaII.png|thumb|300px|center|Figure 1: This assay compares the enzymes present in the regitry i.e, BglII and BamHI to the enzymes added by us, S7 and CviAII. This shows that S7 and CviAII degrade the DNA much quicker than BglII and BamHI combined.]]<html> | ||
| + | |||
| + | <h2>Week 22 (October 1 to Oct 3)</h2> | ||
| + | |||
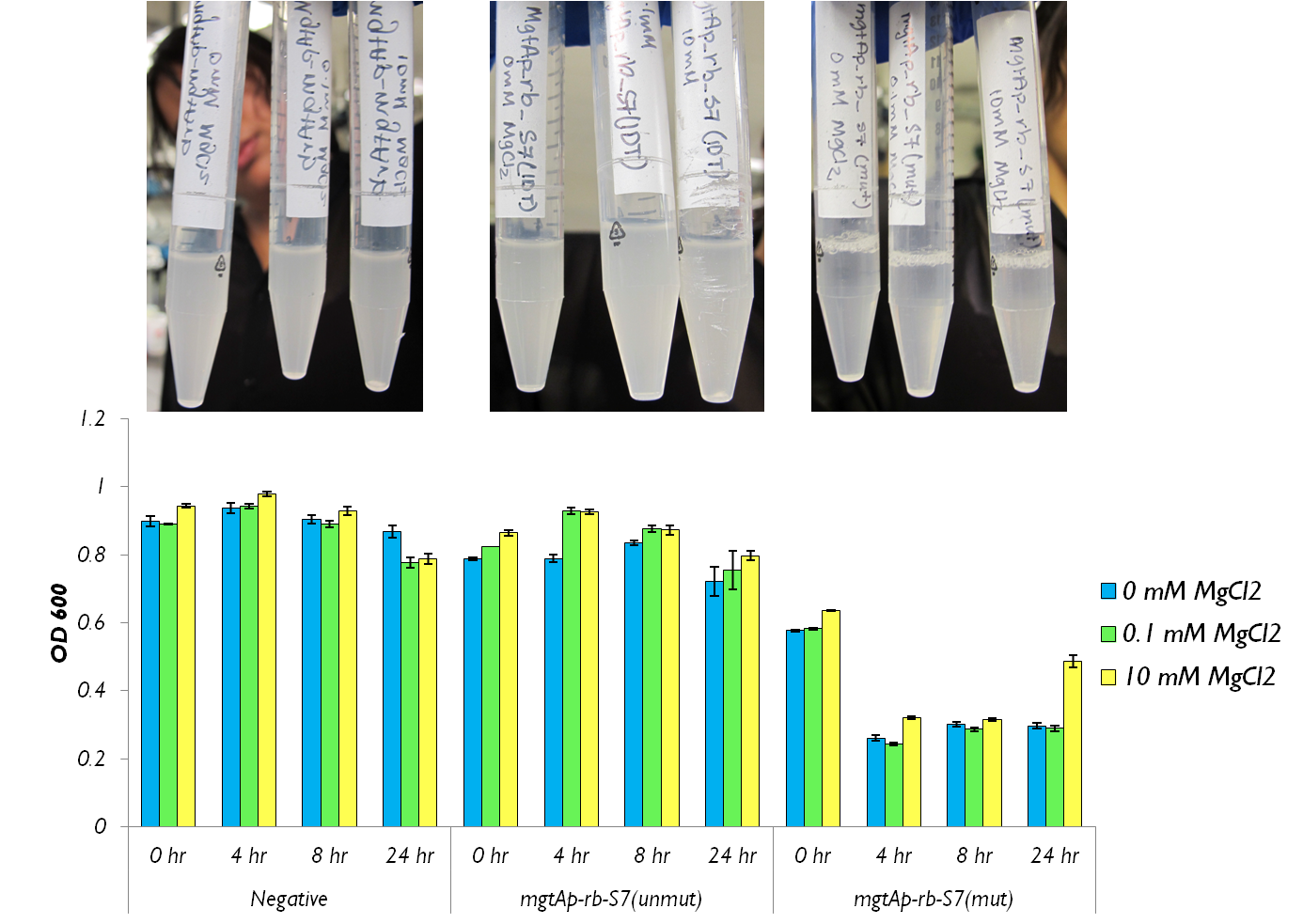
| + | <p> The molybdate assay was attempted this week. mgtAp-rb-S7 construct was tested with mutated S7 and the synthesized S7. OD readings and CFU measurements were taken. </html>[[File:24 hour assay with mgtap-rb-S7 Ucalgary.png|thumb|750px|center| Figure 2: This shows the OD600 values of mgtA circuits with S7 both mutated and unmutated. The negative control consists of <i>mgtAp-mgtArb</i>.]]<html> | ||
| + | <p> Figure 2 shows that the mgtAp-mgtArb-S7 (<a href="http://partsregistry.org/wiki/index.php/Part:BBa_K902018">BBa_K902018</a>) starts acting approximately 4 hours after induction. However,it also shows that 10mM MgCl<sub>2</sub> is not enough salt to inhibit the entire system because there is no difference in OD600 measurement at 4hr timepoint between 10mM and the 0mM concentrations. This test needs to be repeated with higher concentrations of Mg<sup>2+</sup> however this data suggests that the mutagenesis was successful and S7 is active and killing the cells at approximately 4hr which does not necessarily reflect upon the activity of S7 but also reflects upon the response time of the mgtA system.</p> | ||
| + | </p> | ||
| + | |||
| + | <h2>Week 23 (October 9 to October 12)</h2> | ||
| + | |||
| + | <p> The molybdate assay was not successful as the M9 media was contaminated. The assay was deemed not priority. Constructions of constitutive promoter and riboswitch were attempted with GFP (LVA tag) along with other constructions to complete the circuits shown in the regulation page. The constructs were transformed. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | With respect to the rhamnose expression system, our team repeated constructions of the following: Prha-B0034-GFP; and R0040-B0034-rhaS. However, given the trouble we had constructing these circuits, I decided to use gel extraction to isolate purified insert and vector following cutting with appropriate restriction enzymes. As well, I ensured that inserts were in an exact three to one ratio to their respective vectors. Finally, 1 microlitre of a 100mM solution of ATP were added to all ligation reactions since we suspected ATP degradation in our ligase buffer. The ligation reactions were carried out at room temperature overnight, and were frozen for the weekend of Regionals. | ||
| + | </p> | ||
| + | |||
| + | <h2> October 12 to October 14: iGEM Regionals!</h2> | ||
| + | |||
| + | <h2>Week 24 (October 15 to October 19)</h2> | ||
| + | <p> With MOCO, the transformants which grew were amplified using colony PCR. When the samples were run on a gel the bands did not line up properly. Sequencing results came back and TetR promoter with Moco riboswitch was sequenced verified as well as the moco riboswitch with kill gene S7. Further constructions to complete the circuit were attempted and we ended up with 2 sequenced verified constructs of TetR promoter with moco riboswitch and S7 and GFP respectively. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | With the rhamnose system, the ATP spiked ligation reactions of Prha—B0034--GFP and R0040—B0034--rhaS were transformed into E. coli—a number of colonies were produced which were subsequently analyzed with PCR and digestion of isolated plasmids. There were colonies in both construction that appeared to be of appropriate size. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | We are hoping to complete the final rhamnose characterization circuit (Prha—B0034--GFP--R0040—B0034—rhaS) prior to finals; to this end, I designed primers for the <a href="https://2012.igem.org/Team:Calgary/Notebook/Protocols/Gibson_Assembly">Gibson assembly</a> of this circuit from plasmid stocks of pRha, B0034—GFP, B0034—rhaS, and pSB1C3. | ||
| + | </p> | ||
| + | |||
| + | <h2>Week 25 (October 22 to October 26)</h2> | ||
| + | <p> The moco circuits were further constructed with a LacI promoter, a ribosome binding site, and the moaABCDE operon. This was sent to the registry. Characterization of these circuits are in progress. </p> | ||
| + | |||
| + | <p> | ||
| + | With the rhamnose system, sequencing results showed that we successfully constructed Prha—B0034—GFP. There was a mistake in the order so that R0040—B0034--rhaS was not send for sequencing. We will attempt to assemble the final kill switch construct using Biobrick Standard methods. Additionally, we will conduct an assay of Prha—B0034—GFP to see how rhamnose, glucose, and lack of these sugars might affect its fluorescence. The protocol can be seen <a href="https://2012.igem.org/Team:Calgary/Notebook/Protocols/Prha_Characterization">here</a>. We successfully show induction of this promoter with rhamnose and repression with glucose. See the <a href="https://2012.igem.org/Team:Calgary/Project/HumanPractices/Killswitch/Regulation">results</a> on our kill switch page. | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | Also with rhamnose, we attempted to use Pfu to PCR out four elements for the Gibson assembly of the final rhamnose construct. The reactions were conducted on a 50 degree Celsius to 64 degree Celsius temperature gradient. Two of the PCR components were successfully amplified (B0034—GFP and pSB1C3); the other two elements require further optimization (B0034-RhaS and Prha). | ||
| + | </p> | ||
| + | |||
| + | </html> | ||
| + | }} | ||
Latest revision as of 03:23, 27 October 2012


Hello! iGEM Calgary's wiki functions best with Javascript enabled, especially for mobile devices. We recommend that you enable Javascript on your device for the best wiki-viewing experience. Thanks!
Team Killswitch
Week 1 (May 1-4)
This was the first week where we met with other team members and summarized the primary subprojects the team will be tackling this coming summer.
Week 2 (May 7-11)
Members were assigned to the killswitch team. We spent a majority of this week performing literature searches and narrowing the killswitch to a few ideas.
The mechanism of death is still settled upon the micrococcal nuclease, but regulation of the death genes will be difficult. Existing repressible promoters in the registry still tend to be leaky in their expression, but we need to test this out. We may look into the TetR-repressible promoter (R0040) and also the clλ regulated promoter (R0051).
We also found a few riboswitches responsible to various metal ions such as Mg2+ and Mn2+. The mgtA riboswitch activates translation of the death gene when magnesium ions are not present in the solution. The mntA riboswitch deactivates translation of the death gene when manganese ions are not present. Considering this, we may be able to find a method of precipitating or otherwise sequestering Mn2+ ions out of the tailings water prior to entering the bioreactor. This way, our bacteria would not die when they come into contact with the manganese in the tailings water. Possible additives include carbonate (CO32-) or hydroxide (OH-), though this may alter the pH greatly.
One other proposal would be to use promoters that activate when the bacteria detect the formation of a biofilm, for example, on glass beads. Engineering a monolayer of cells on a bead means that if a cell detaches, its death genes will activate. Further research into this is needed.
Week 3 (May 14-18)
We are continuing to look in to various other pathways. We discussed the possibility of a NOR-gated system to add a second layer of regulation to the kill gene. It would require the production of riboswitch ligands. Possible ligands include glucose, amino acids, and molybdenum cofactor.
We also looked into a co-dependent system where we would transform E. coli with two different plasmids and put them in the same bioreactor such that they produce ligands for each other that would repress the expression of the restriction enzyme and the nuclease. For this we explored riboswitches further and came up with the idea of using MOCO and SAM riboswitches as SAM is soluble in water and not found in the tailings ponds.
We also found a glucose repressible promoter that we could have potentially used upstream of the kill switch. This promoter is found in F. tularensis. However we also found that this promoter is not found in E. coli, and F. tularensis has a unique polymerase which used this promoter. Hence using this promoter does not seem feasible. Next week we will be looking into further possibilities for the kill switch.
Week 4 (May 21-25)
In terms of our literature search we decided against the glucose repressible promoter from F. tularensis because it was not native to E.coli and upon contact with the authors of the paper, we found that the promoter did not work in E.coli. We also came upon two systems that we are interested in further investigating. The first one was a glucose activated promoter that was found in the registry which we are thinking of coupling to an inverter and the second one is a rhamnose-based regulatory system.
On Tuesday, in addition to our literature research, we decided on some of the circuits that we will be building over the next couple of weeks and came up with a plan detailing what will be done each day. These are the circuits:
On Wednesday we proceeded with the transformation of 3 parts from the iGEM kit into our E.coli: a tetR promoter, a GFP-double terminator, and a RBS-GFP-double terminator. On Thursday we verified our transformation process with a cPCR where we ran 10 colonies from each plate with our controls. We ran a gel of the PCR products and viewed it to verify the transformation and to decide which of the successful colonies to plate and grow overnight.
On Friday we isolated the plasmids through a mini prep. The plan for next week is to do a restriction digest and run it on a gel to confirm that the plasmids have the part. We also want to start more transformation of our parts into E.coli such as the riboswitches and the promoters. We want to verify the transformation, isolate the plasmids and verify, and start the DNA construction digest to start building our circuit.
Week 5 (May 28-June 1)
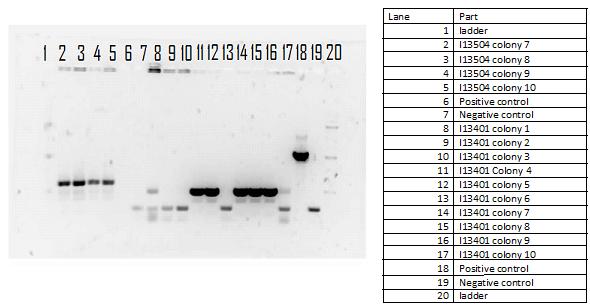
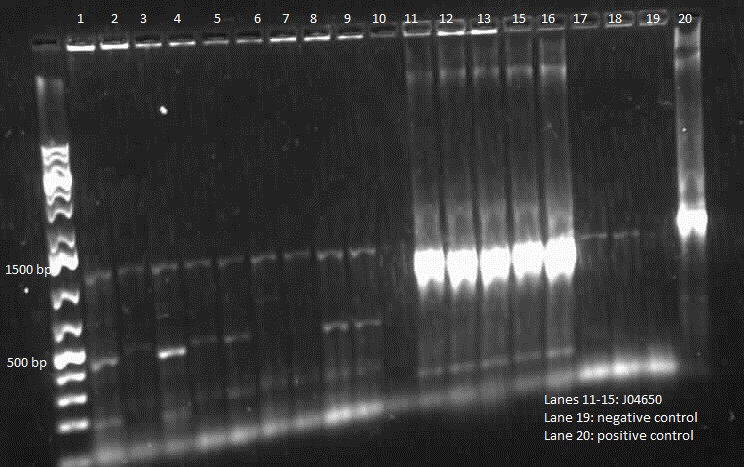
During the week of May 28th, the killswitch team made progress toward transforming/verifying nine different parts in E. coli. Work began with continuation of the previous week's transformation of R0040 (TetR promoter), I13401 (GFP terminator), and I13504 (RBS GFP terminator). Note that these three parts will be necessary for characterization of the four different kill system triggers being explored by the killswitch team. Verification procedures (ie, colony PCR) showed successful transformation of I13401. Results for the other two parts were inconclusive and cloning of R0040 and I13504 will continue next week.
The commercially synthesized mntA promoter, mgtA promoter, and mgtA riboswitch for the magnesium and manganese riboswitches were also transformed into E. coli. While colonies were successfully isolated, colony PCR was inconclusive because incorrect primers were used for the commercial plasmids. The team proceeded to isolate the plasmids from E. coli, perform a restriction digest of the parts, and ligation of these components into biobrick vectors. These products were transformed into other E. coli and the team is ready to verify presence of the cloned digest products.
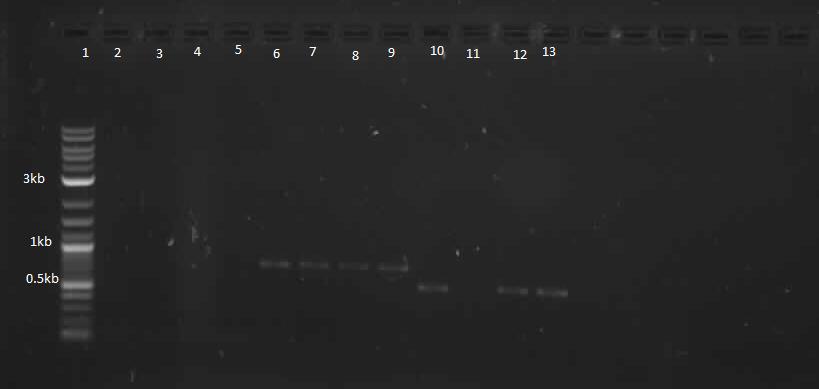
In continuation with magnesium and manganese riboswitches, the team attempted to biobrick mntR (a repressor of a manganese transporter activated by manganese), PhoP (a transcriptional regulator of mgtA), and PhoQ (a signal transducer which activated PhoP in the presence of divalent cations). The below PCR indicates that while PhoQ and mntR were successfully isolated, PhoP was not. The former two were transformed into E. coli; the transformed samples grew into a lawn and colony PCR will thus be performed next with re-streaked plates. Finally, gradient PCR was used in an attempt to biobrick PhoP.
Lanes 6-9: PhoQ (~671 bp)
Lanes 10-13: mntR (~500 bp)
Primers for the MOCO and rhamnose killswitch actuators were also designed. With respect to the MOCO synthesis operon and MOCO riboswitch, primers were finalized and sent to senior members of the team. While primers for isolation of rhaSR and the rhaT promoter from E. coli were given in the paper which inspired the rhamnose promoter idea, the team was resistant to using the sequences since they did not seem to correlate with genes of interest in a BLAST search. Closer inspection revealed that the forward primer listed in the paper added a number of bases (>50) which were not native to the E. coli genome. Only 18 base pairs of the primer were complementary to E. coli—this discrepancy was responsible for the negative results of the BLAST alignment. Work will continue next week to optimize the rhamnose primers. They hope to order both the MOCO and rhamnose primers next week so that they may proceed with construction of the kill switch.
Week 6 (June 4th-8th)
This week we continued our attempt to biobrick PhoP, PhoQ and mntR. A gradient PCR was run with temperature and salt gradient (0.5mM-3.0mM). however this PCR showed no success whatsoever. We discovered that the primers designed for these parts were faulty and were binding in several places in the gene and thereby favouring products that are smaller in size.
phoP-S GTTTCTTCGAATTCGCGGCCGCTTCTAGatgCGCGTACTGGTTGTTG
phoP-AS GTTTCTTCCTGCAGCGGCCGCTACTAGTATTATTAtcaGCGCAATTCGAACAGATAG
phoQ-S GTTTCTTCGAATTCGCGGCCGCTTCTAGatgaaaaaattactgcgtc
phoQ-AS GTTTCTTCCTGCAGCGGCCGCTACTAGTATTATTAttattcatctttcggcgcag
mntR-S GTTTCTTCGAATTCGCGGCCGCTTCTAGatgagtcgtcgcgcaggtac
mntR-AS GTTTCTTCCTGCAGCGGCCGCTACTAGTATTATTAtcatttggcaccgtgtttc
Hence new primers were designed.
PhoPNEW-AS CTGCAGCGGCCGCTACTAGTAGCAGTAATTTTTTCATCAGCGCAATTCG
PhoQNEW-AS CTGCAGCGGCCGCTACTAGTAGCATATTTATTCATCTTTCGG
mntRNEW-S gaattcgcggccgcttctagGAGGAAGCACAATGAGTC
mntRNEW-AS CTGCAGCGGCCGCTACTAGTAGCGTGCGTAAAAAAGG
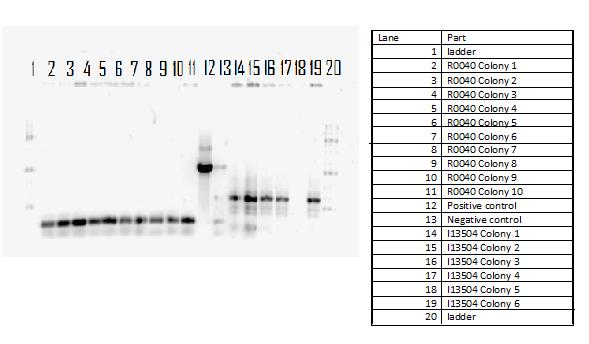
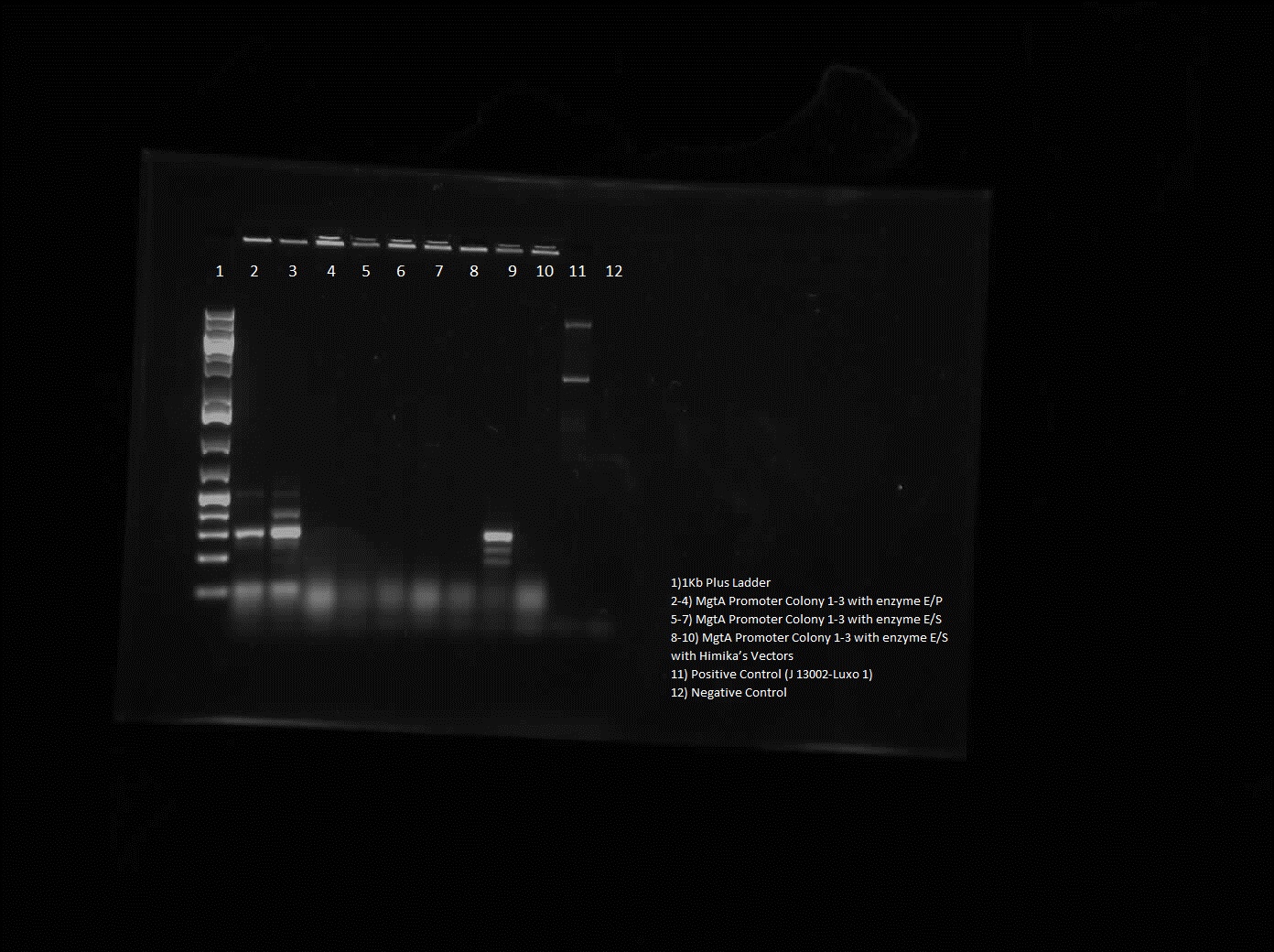
The killswitch team also focused on verification of both the magnesium/manganese (MgtA promoter, MgtA riboswitch and MntP promoter) and registry parts (R0040, I13401 and I13504).
For the magnesium/manganese parts this week we insert them in biobrick vectors in order to carry out colony PRR. Colony PCR of the magnesium/manganese parts could not have been done first since we did not have the right primer , therefore by doing a construction digest we could use the standard biobrick primers. From the gel, the results were inconclusive as the ladder was not clear and the parts were too small to tell the difference between them and primer dimers, therefore next week goal is to perform another CPCR of the parts. As well as doing colony PCR of the MgtA promoter, a restriction digest of this part was done. The gel showed good results as the bands were bright and the right size.
For the registry parts R0040 no results were shown in the initial colony PCR, however another gel was done with R0040 from the 2010 and 2011 registry plate. The colony PCR of R0040 from 2011 showed the rights bands, however the results are inconclusive as the negative was contaminated, therefore most likely another colony PCR needs to be done.
The registry part I13401 had a successful week as both the colony PCR and the restriction digest verified our parts. Since the result was successful we performed glycerol stocks of the plasmid and prepared it for sequencing the following week.
The part I13504 showed no results this week therefore the goal next week is to find another part that is similar to I13504 and test its verification.
Week 7 (June 11-14)
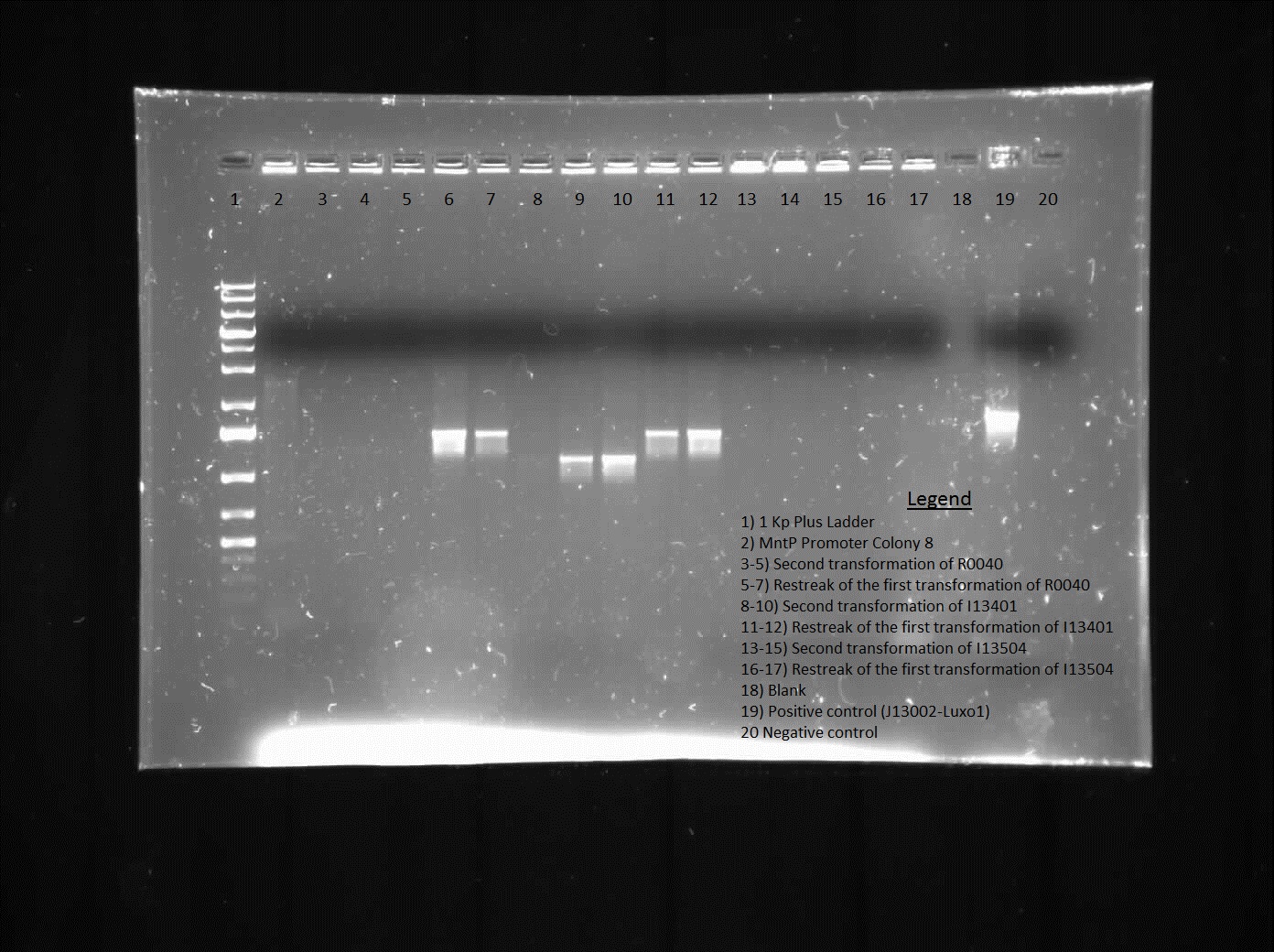
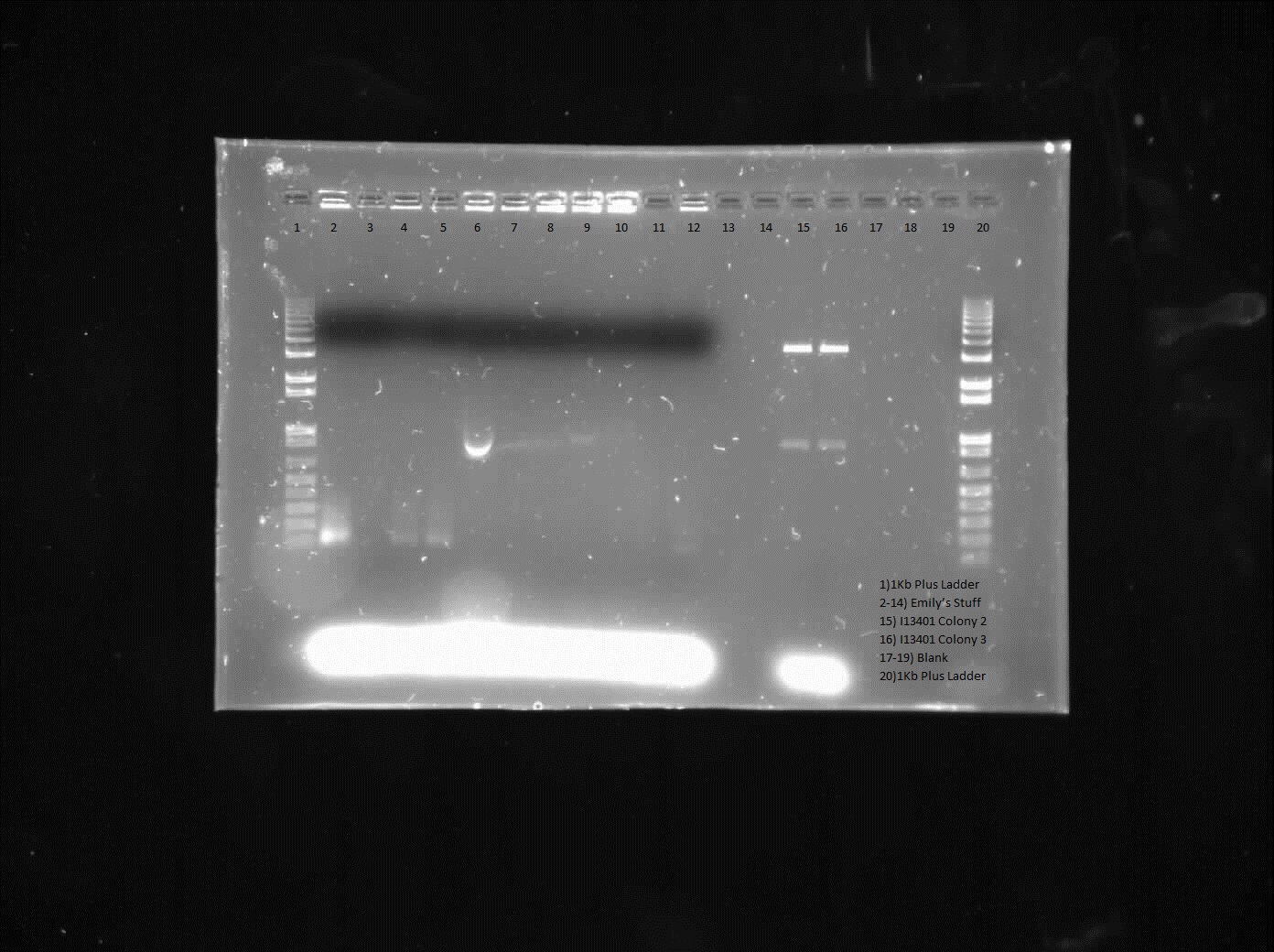
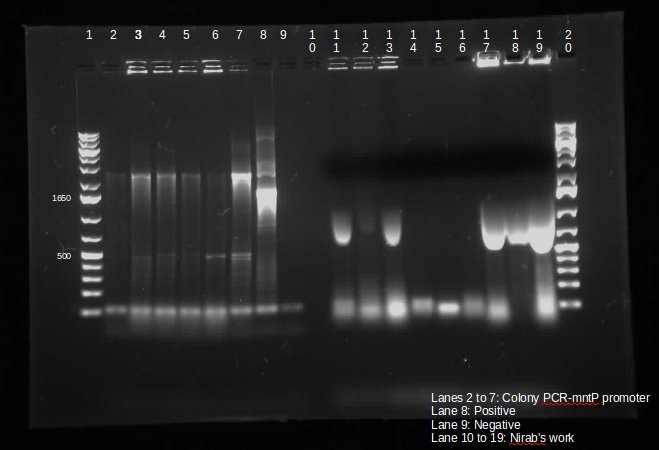
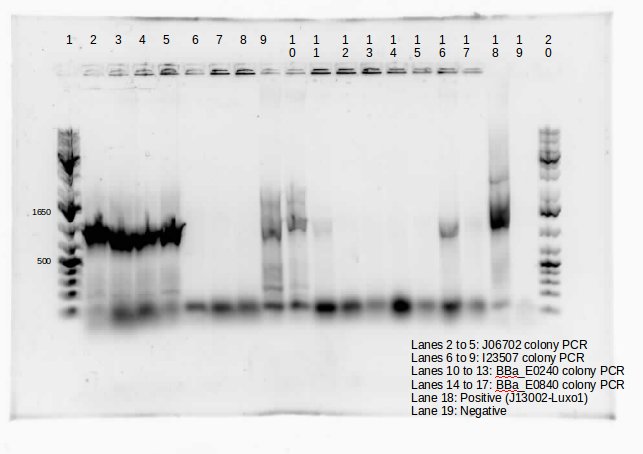
This week we continued the insertion of magnesium promoter (MgtA) into biobrick plasmid (PSB1C3) to carry out the colony PCR to confirm the presence of the Mgta part in the plasmid using Bbk_CP_F and Bbk_CP_R primers. We are currently in the process of further verification by using a mini prep. We also transformed four registry parts (J06702 [RBS-mCherry-dTerm], I13507 [RBS-RFP-dTerm], E0840 [RBS-GFP-dTerm] and E0240 [RBS-GFP-dTerm]) directly from the kit plates and verified the parts using colony PCR the following day. The primers used to verify these parts were Bbk_CP_F and Bbk_CP_R. These parts were successfully transformed but we still saw some contaminant bands in the gels (see below).
]
Transformation of MntP in PSB1C3 vector was verified using colony PCR. However, the results indicated that the presence of MntP as well as a contaminant band around 3000 bp.
]Transformation of registry parts I13521 [TetR-B0034-RFP-dTerm], I13507 and J04650 [RFP-dTerm] were performed and colony PCR was carried out to verify the presence of the parts in the cells. J04650 was the only part that was successfully transformed. Though the other parts were not successfully transformed, we can still see non-specific bands in the gel (see below). We are currently doing mini prep to additionally verify the success of transformation.
Week 8 (June 18-June 22)
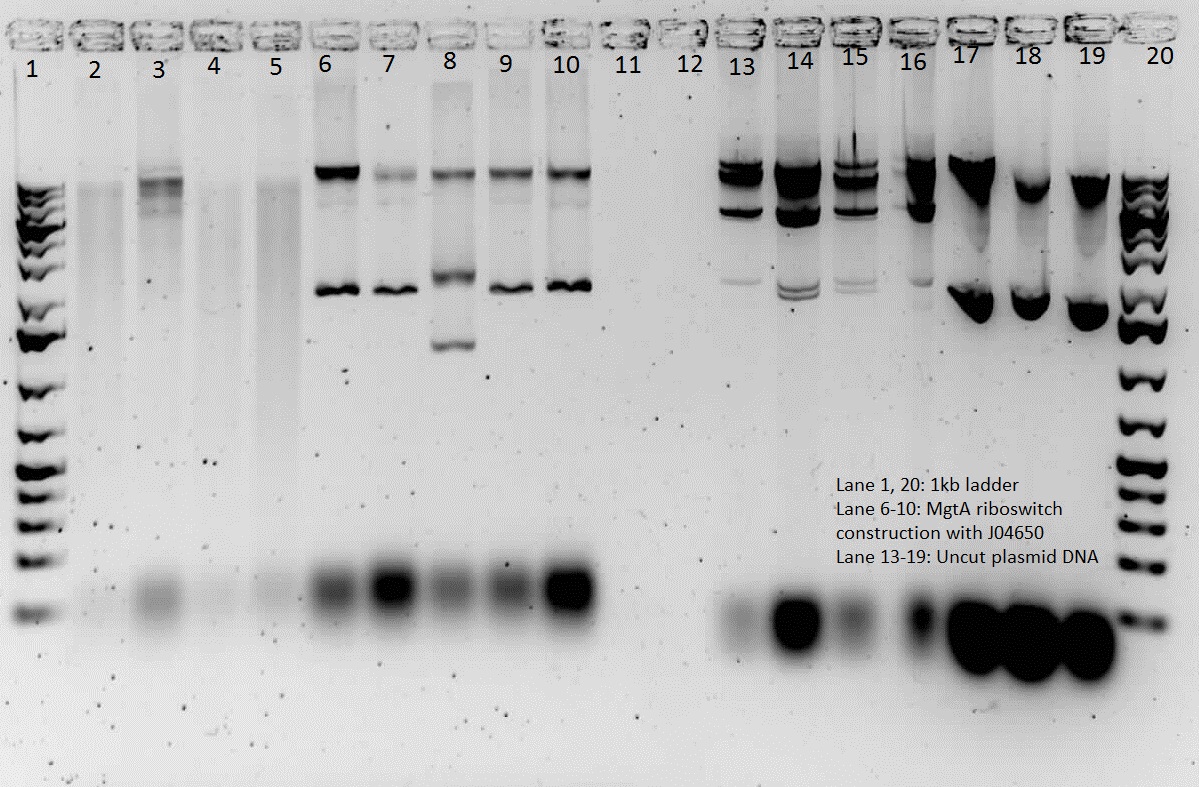
This week we did a restriction digest of all the plasmids that were isolated. We confirmed that we had J04650, I13507, E0240 and mgtA riboswitch. Hence these were sent to sequencing. We got sequencing results back and confirmed that we have J04650, I13507, J06702, R0040, E0240 and mgtA riboswitch. Henceforth, we started constructing the mgtA riboswitch with J04650.
The mgtA riboswitch with J04650 was confirmed using PCR as well as restriction digest. We plan on sequencing it when we are able to construct it with a promoter.
Week 9 (June 25-June 29)
This week we attempted to construct the mgtA riboswith and J04650 construct with promoters such as MgtA and TetR. We also tried to plasmid switch I13507 such that we can put mgtA promoter on to that composite part. So far we have verified through colony PCR that it worked. This was verified again with restriction digest however the gel did not look too good therefore we ran the inserts and vectors to see how they looked. We will reattempt this again next week.
Week 10 (July 3-July 6)
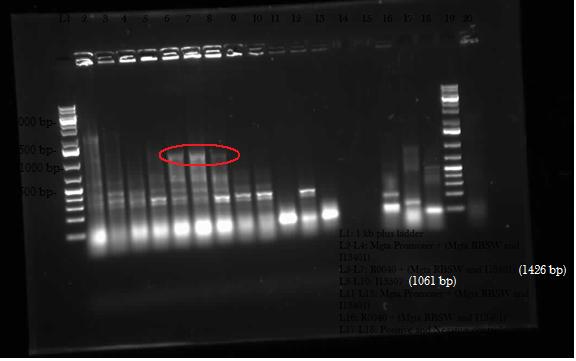
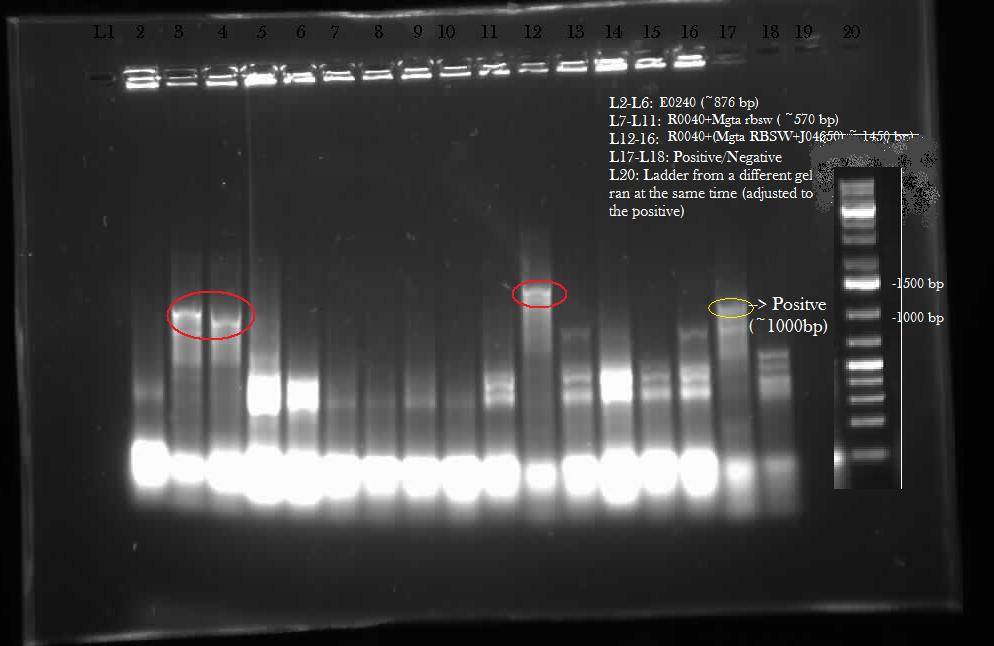
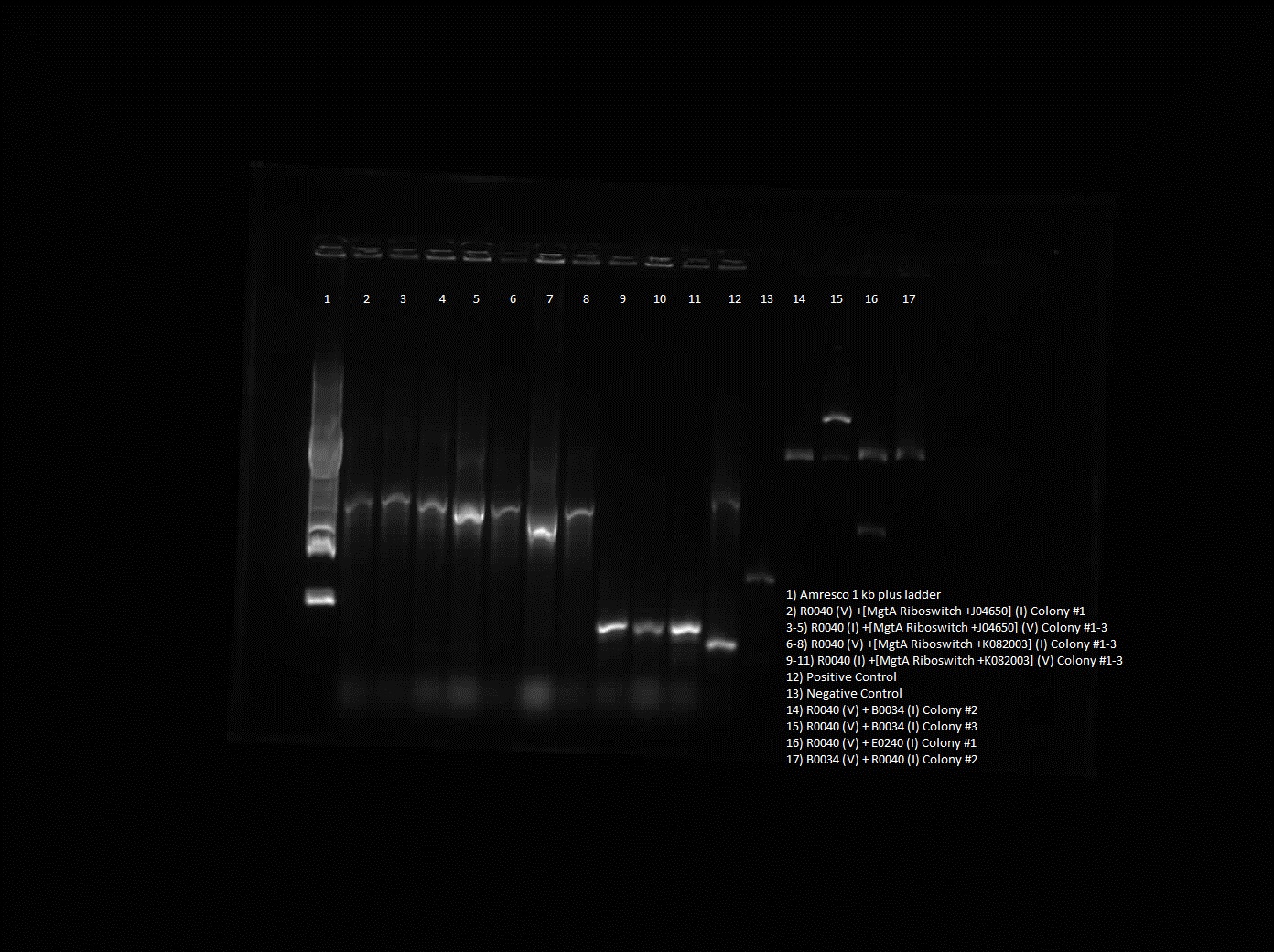
This week, we attempted to religate our constructions from the previous week as they were not verified as positive in our colony PCR. We had run a gel to see if the inserts and vectors were digested properly and they were. Following transformation, we verified it through cPCR. The two gels show the results of construction involving two promoters (Mgta Promoter and a constitutive tetR promoter, R0040) and the mgta rbsw with different reporter gene constructs from the previous week. There are also some plasmid switches and constructions of the promoters and just the riboswitch. Based on these results, we isolated plasmids of R0040 + (Mgta RBSW and I13401) from L6 and L7 as well as I13507 from L8 from the first gel and R0040 + (Mgta RBSW and J04650) from the second gel (circled in red). We also transformed B0034, a ribosome binding site and constructed Mgta rbsw with a reporter construct with fast degrading GFP.
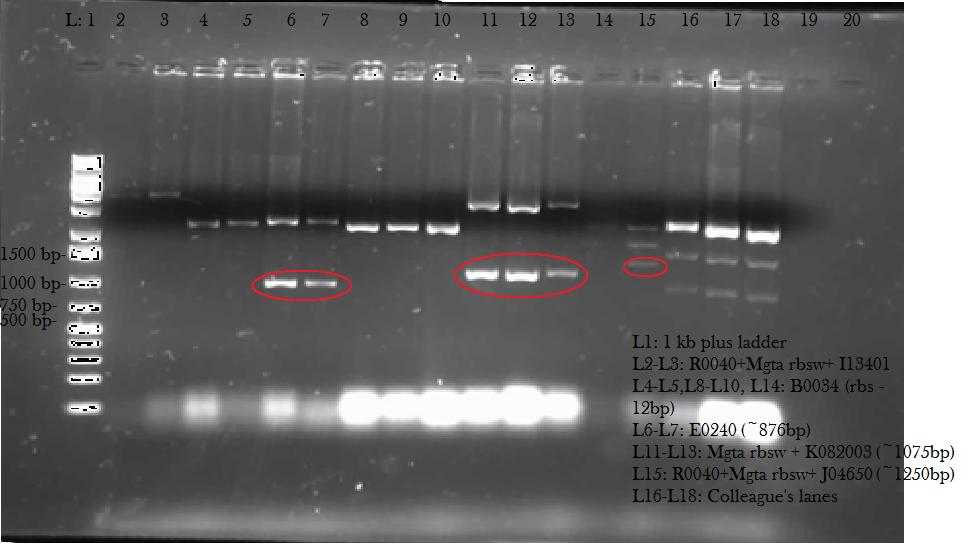
We verified these constructions with a digest. The third gel shows the digest and the lanes circled are the ones that look good and were sent to sequencing.
Week 11 (July 9-July 13)
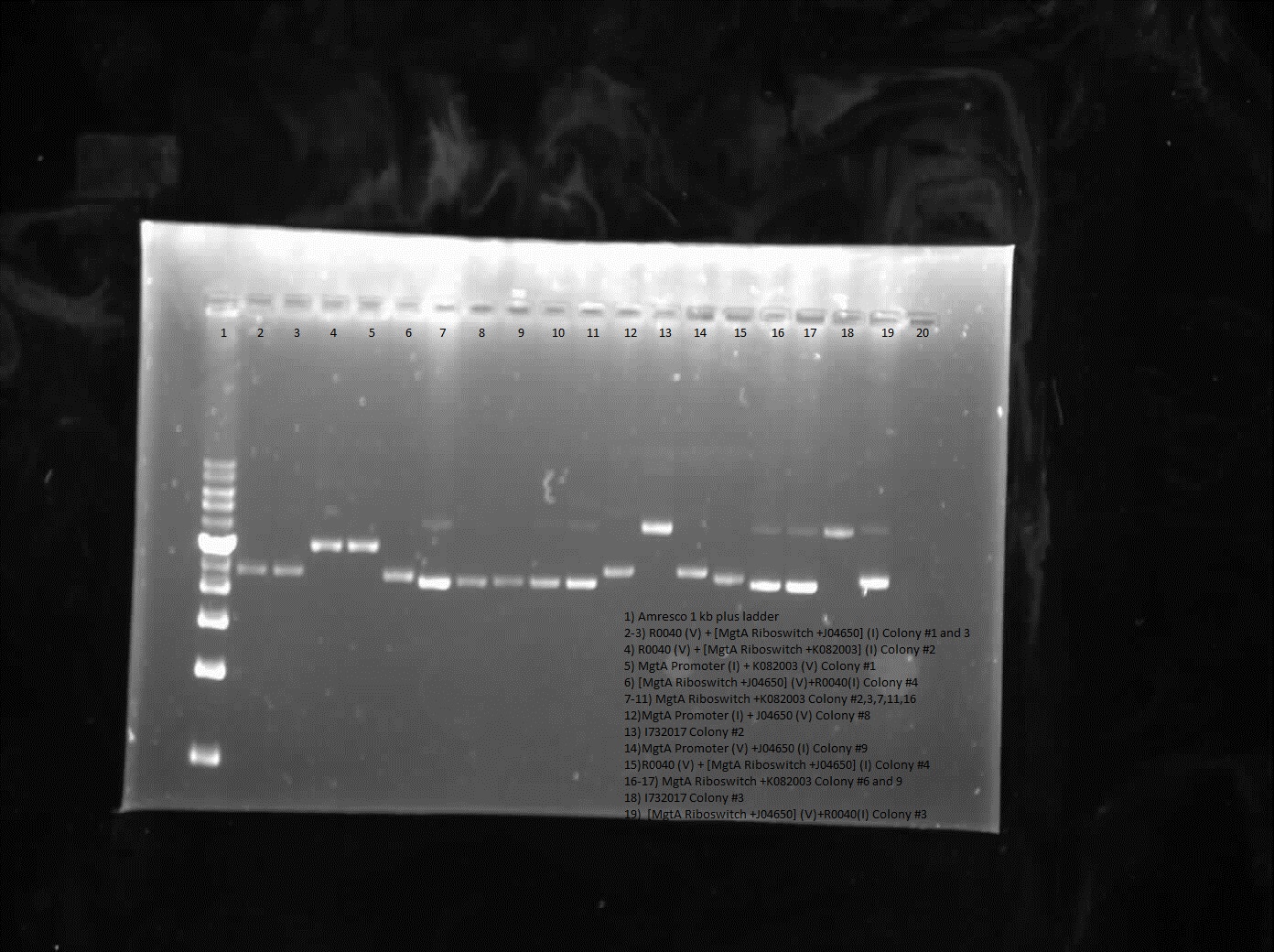
During the week of July 9th, construction was done on R0040 with J04650 and K082003 constructs as well as E0240 and B0034. Similarly the MgtA promoter was constructed with either J04650 construct, K082003 construct, E0240 or B0034. Furthermore a gel was run on the J04650 and K082003 constructs to check for genomic contamination which showed positive results for sequencing.
From the restriction digest of the construction R0040 or MgtA promoter with the J04650 or K082003 constructs little results was shown, however some of the sequencing presented varying results. Sequencing of the R0040 with MgtA riboswitch and J04650 worked beside for weird sequencing around the scar site while both registry part E0240 and B0034 were good. The construct with MgtA promoter did not work as sequencing showed that the promoter was not part of the construct therefore the construction of these parts will have to continue another week.
Results from colony PCR showed positive bands for R0040 (vector) with B0034 (insert), R0040 (vector) with E0240 (insert) and B0034 (vector) with R0040 (insert).
Construction of these constructs will be done another week as the restriction digest did not look good at the same time the ladder was not readable.
Week 12 (July 16 to July 20)
During week twelve (July 16th to July 20th), the kill switch team worked on three different projects: first, transformation/verification of kill genes; second, transformation and PCR of parts for the rhamnose inducible kill system; and third, construction of parts for characterization of the magnesium riboswitch kill system.
The commercially synthesized kill genes K131009 (colicin E2 operon w/o SOS promoter) and S7 nuclease (micrococal nuclease gene) were transformed into E. coli. Colonies transformed with these genes, along with the previously transformed K117000 (for colicin activation), K112808 (holin-antiholin lysis system), and endolysin, were subjected to verification procedures including colony PCR and verification digests. Successful transformants were sent off for commercial sequencing.
The team also began work on the the rhamnose inducible kill system since primers for rhaR and rhaS genes were received along with the synthesized rhamnose promoter. Temperature gradient PCRs (54C-59C; also 54C-65C with phusion)for the rhaR and rhaS genes were performed with no success. The rhamnose promoter was transformed into E. coli of which a plasmid stock was isolated by miniprep.
Finally, the team continued with constructions necessary for characterizing the magnesium riboswitch kill system. They continued with verification of the MgtA riboswithch with GFP and also GFP(+LVA tag) under a constitutive promoter.
Week 13 (July 23 to July 27)
During the week thirteen, the killswitch team performed plasmid switches of S7, mntP riboswitch and pRha (rhamnose inducible promoter) from the commercial IDT synthesis vector into pSB1C3.
Additionally, the team attempted to remove RhaR and RhaS from the E. coli genome via PCR. Kappa polymerase was used with a 50-65 degrees Celsius temperature gradient with no success. Given this failure, touchdown PCR from 58 to 74 degrees Celsius was performed on the RhaR and RhaS genes. These strategies failed. The team re-analyzed the primers used in these protocols; the antisense primers were erroneous and had to be redesigned.
Additionally, we tried construction of mgtAp with mgtArbsw-K082003. However no positive clones appeared in the follow up verification.
We also worked on construction of R0040+B0034, R0040+E0240, R0040+MgtA Riboswitch, R0011+MgtA Riboswitch, MgtA Promoter+B0034, MgtA Promoter with MgtA Riboswich and GFP(+LVA tag), and B0034 with GFP(+LVA tag). These constructions are all in different phases of the verification process. The team also attempted to expedite construction of pRha (rhamnose promoter) and S7 (micrococcal nuclease) with BOO34 by building the directly from the commercial IDT synthesis vectors; however, this process failed due similar resistance genes on the IDT and BOO34 vectors.
Week 14 (July 30 to August 3)
This week we tried to construct R0040 and R0011 with mgtArb-K082003. This construction was done both ways, i.e both R0040 and mgtArb-K082003 were used as both vectors and insert. Out of these 2 constructions, R0040-mgtArb-K082003 gave positive clones however R0011-mgtArb-K082003 did not. A sample from a single colony with R0040-mgtArb-K082003 in pSB1C3 was sent to sequencing; the results were postively verified.
Additional constructions that were started include the following:
R0011-mgtArbs
R0011-mgtArbs-K082003
mgtArb-S7
pRHA-B0034
B0034-S7
R0011-B0034
mntPrbs-K082003
mntPrbs-S7
B0034-S7 (ie, S7 in pSB1C3)
Digest verification showed success with K082003+B0034, J13002+K082003, R0040+ MgtA Riboswitch, and R0040+E0240; henceforth these were submitted for sequencing.
Week 15 (August 6 to August 10)
Colony PCR was performed on constructions from the previous week. Positive results were obtained with the following constructions: R0011-MgtAriboswitch, MgtAriboswitch-S7, MntpRiboswithch-S7, R0011-B0034, and MntpRiboswich-K082003. Minipreps and verification digests were performed from the relevant colonies.
The commercially synthesized Mntp riboswitch and moco riboswitch were switched into PSB1C3 vectors. Colony PCR and restriction digest verification yielded positive results; these samples are destined for sequencing verification. Additionally, the team constructed tetR-RBS-GFP(+LVA. Verification procedures were performed. TetR-RBS-GFP(+LVA) is awaiting sequence verification.
Temperature gradient PCR and salt gradient PCR were used to optimize and amplify the moa operon from the E. coli genome. The rhaR gene was amplified from Top Ten E. coli colonies and and biobricked in pSB1C3 with Kapa polymerase. The rhaS gene was amplified from Top Ten E. coli on a 52 to 67 degree Celsius temperature with Kapa polymerase and DMSO.
Continued working with MgtA Promoter+ B0034, R0011 +MgtA Riboswitch,B0034+K082003 and trying to get DNA of the MntP Promoter.
Week 16 (August 13 to August 17)
For the week of August 13th continuation of the construction of the MgtA Promoter +B0034, R0011+MgtA riboswitch and B0034+K082003 was conducted. Constructions of MgtA Promoter+B0034 and B0034+K082003 were tested with plasmid PCR several times however none of the plasmid PCR showed any bands on the gel. R0011+MgtA riboswitch restriction digest did not work therefore a reconstruction was done and will follow thought for the next week. One of the colonies from MntP Promoter IDT plate digested well and was therefore plasmid switch into pSB1C3.
We started plasmid switching parts that were not in pSB1C3 into pSB1C3. Additionally, we also started to characterize the magnesium system using mgtAp-mgtArb-K082003, mgtAp-mgtArb, R0040-mgtArb-K082003. We also tried to construct R0011-mgtArb, mntPrb-S7, mntPrb-S7J13002-K082003, R0011-mgtArb-K082003, B0034-S7. At the end of the week we were able to sequence verify B0034-S7 and mntPrb-K082003.
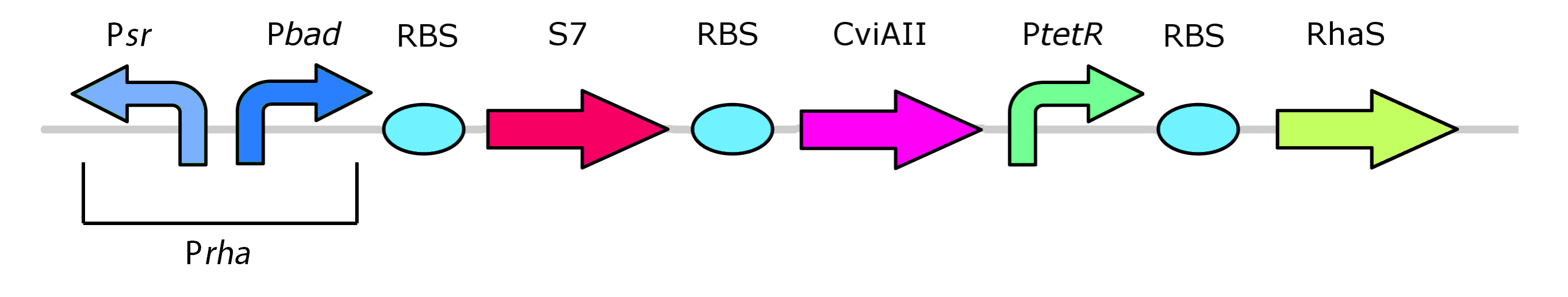
Finally, the rhamnose promoter system has been reoriented:
In this finalized version of the construct, the RhaR gene has been left out from the system. Given that RhaR activates RhaS, and RhaS activates the pRha promoter, it should theoretically be sufficient to constituitively express the RhaS promoter to activate the system. Construct must continue so that it can be tested whether this new design will function and be repressible with glucose. Check out the project page for more details.
Week 17 (August 20 to August 24)
For the week of August 20th most of the constructions were redone while we continue with any other constructions we have already started since we noticed that we haven’t fully heat kill the enzymes. MgtA Promoter + B0034 was constructed with another B0034 that was in a kan vector while the MntP Promoter was redone from IDT as the previous MntP Promoter was not working.
This week we tried to construct mgtAp-mgtArb-S7-mgtArb so that we can construct CviAII when it would arrive.
We also attempted redoing the nuclease assay with S7 and E. coli genomic prep.This time the three different amounts of S7 was used. We used 200U, 50U and 10U.
The moa operon was biobricked and sequence verified. We are trying to construct the moa operon with a costitutive promoter and a ribosome binding site.
Constructions of the rhamnose promoter (pRha) with GFP and S7 continued. These constructions have proven challenging and will have to be repeated next week.
Week 18 (August 27 to August 31)
For the week of August 27th constructions of MgtA Promoter +B0034, MntP Promoter +B0034 in Kan, R0010+ MgtA Riboswitch, R0010 +MntP Rbsw + S7, MntP promoter + MntP Riboswitch + S7 had to be redone as the ones done on last Friday did not grow or verification through PCR did not show positive results. After repeat trails of these construction CPCR showed that positive bands for MntP Promoter plasmid switch into kan vector, MgtA Promoter+B0034, MntP Promoter +B0034 (Kan), R0010 +MgtA Riboswitch and R0010 + MntP Riboswitch + S7. Since there was positive results was shown for these constructions, mini prep and digestion verification will be done the following week.
This week we kept reattempting our constructions however sequencing came back as things it should not have been randomly. Mostly Car genes from the denitrogenation team. Therefore we had to back to our glycerol stocks from the previously verified part and try to reconstruct our parts.
Week 19 (September 3 to September 7)
For the week of September 3rd constructions (MntP Promoter plasmid switch into kan vector, MgtA Promoter+B0034, MntP Promoter +B0034 (Kan), R0010 +MgtA Riboswitch and R0010 + MntP Riboswithc + S7), was verified with restriction digest and sent to sequencing. Also MntP Promoter+ MntP Riboswitch +S7 construction was reattempted after the enzyme incident where they were not fully heat killed.
Week 20 (September 10 to September 14)
The moco riboswitch was biobricked and sequence verified. Constructions of the moco riboswitch with different promoters (R0010, R0011, R0040) were attempted along with the moco riboswitch with the GFP LVA tag, CviaII and S7. The attempts of construction of the moa operon with the constitutive promoter and a ribosome binding site have not been successful thus far and is an ongoing project.
Some of the constructions of pRha and GFP looked promising in colony PCRs and verification digests. These samples were sent for sequencing. In the meantime, we attempted to induce the potential colonies with rhamnose to see if there was evidence of a successful construction prior to sequencing results. Results were negative compared to constituitively expressed GFP and a negative control. We awaiting sequencing results to confirm this failure.
Week 21 (September 17 to September 21)
The constructions with the moco riboswitch and the moa operon were not successful and were reattempted this week.
Rhamnose promoter constructions with GFP were negative as shown in sequencing results. We recommenced with this construction.
Week 22 (September 24 to September 29)
The constructions with the moco riboswitch and the kill genes and the GFP with LVA tag looked promising on the gels and were sent to sequencing. We tried to construct those constructs with a promoter. R0010 with the moco riboswitch and CviaII looked promising on the gels and was also sent to sequencing.
This week we reattempted the nuclease assay to compare BglII, BamHI, S7 and CviAII against each other. This time we used 10U of enzyme in each condition and tested degradation at every 45 minutes.
Week 22 (October 1 to Oct 3)
The molybdate assay was attempted this week. mgtAp-rb-S7 construct was tested with mutated S7 and the synthesized S7. OD readings and CFU measurements were taken.
Figure 2 shows that the mgtAp-mgtArb-S7 (BBa_K902018) starts acting approximately 4 hours after induction. However,it also shows that 10mM MgCl2 is not enough salt to inhibit the entire system because there is no difference in OD600 measurement at 4hr timepoint between 10mM and the 0mM concentrations. This test needs to be repeated with higher concentrations of Mg2+ however this data suggests that the mutagenesis was successful and S7 is active and killing the cells at approximately 4hr which does not necessarily reflect upon the activity of S7 but also reflects upon the response time of the mgtA system.
Week 23 (October 9 to October 12)
The molybdate assay was not successful as the M9 media was contaminated. The assay was deemed not priority. Constructions of constitutive promoter and riboswitch were attempted with GFP (LVA tag) along with other constructions to complete the circuits shown in the regulation page. The constructs were transformed.
With respect to the rhamnose expression system, our team repeated constructions of the following: Prha-B0034-GFP; and R0040-B0034-rhaS. However, given the trouble we had constructing these circuits, I decided to use gel extraction to isolate purified insert and vector following cutting with appropriate restriction enzymes. As well, I ensured that inserts were in an exact three to one ratio to their respective vectors. Finally, 1 microlitre of a 100mM solution of ATP were added to all ligation reactions since we suspected ATP degradation in our ligase buffer. The ligation reactions were carried out at room temperature overnight, and were frozen for the weekend of Regionals.
October 12 to October 14: iGEM Regionals!
Week 24 (October 15 to October 19)
With MOCO, the transformants which grew were amplified using colony PCR. When the samples were run on a gel the bands did not line up properly. Sequencing results came back and TetR promoter with Moco riboswitch was sequenced verified as well as the moco riboswitch with kill gene S7. Further constructions to complete the circuit were attempted and we ended up with 2 sequenced verified constructs of TetR promoter with moco riboswitch and S7 and GFP respectively.
With the rhamnose system, the ATP spiked ligation reactions of Prha—B0034--GFP and R0040—B0034--rhaS were transformed into E. coli—a number of colonies were produced which were subsequently analyzed with PCR and digestion of isolated plasmids. There were colonies in both construction that appeared to be of appropriate size.
We are hoping to complete the final rhamnose characterization circuit (Prha—B0034--GFP--R0040—B0034—rhaS) prior to finals; to this end, I designed primers for the Gibson assembly of this circuit from plasmid stocks of pRha, B0034—GFP, B0034—rhaS, and pSB1C3.
Week 25 (October 22 to October 26)
The moco circuits were further constructed with a LacI promoter, a ribosome binding site, and the moaABCDE operon. This was sent to the registry. Characterization of these circuits are in progress.
With the rhamnose system, sequencing results showed that we successfully constructed Prha—B0034—GFP. There was a mistake in the order so that R0040—B0034--rhaS was not send for sequencing. We will attempt to assemble the final kill switch construct using Biobrick Standard methods. Additionally, we will conduct an assay of Prha—B0034—GFP to see how rhamnose, glucose, and lack of these sugars might affect its fluorescence. The protocol can be seen here. We successfully show induction of this promoter with rhamnose and repression with glucose. See the results on our kill switch page.
Also with rhamnose, we attempted to use Pfu to PCR out four elements for the Gibson assembly of the final rhamnose construct. The reactions were conducted on a 50 degree Celsius to 64 degree Celsius temperature gradient. Two of the PCR components were successfully amplified (B0034—GFP and pSB1C3); the other two elements require further optimization (B0034-RhaS and Prha).
 "
"