Team:Calgary/Notebook/PromoterScreen
From 2012.igem.org
| (8 intermediate revisions not shown) | |||
| Line 240: | Line 240: | ||
Genomes of 66-1 and 170-1 were isolated, digested with BglII and with XbaI, and religated before being transformed into <i>E.coli</i>. Cells were plated onto tetracycline plates to isolate cells containing a ligation product with the transposon present. These colonies have been miniprepped, and are awaiting being sent for sequencing to determine which genes the transposon has been inserted into. </p> | Genomes of 66-1 and 170-1 were isolated, digested with BglII and with XbaI, and religated before being transformed into <i>E.coli</i>. Cells were plated onto tetracycline plates to isolate cells containing a ligation product with the transposon present. These colonies have been miniprepped, and are awaiting being sent for sequencing to determine which genes the transposon has been inserted into. </p> | ||
| + | <h2>Week 22 (October 1-October 5)</h2><p> | ||
| + | Minipreps from previous weeks of the transposon self-cloned plasmid were nanodropped, and it was determined that no DNA had been isolated. Therefore, colonies were re-prepped, and sent for sequencing (as sequencing primers have arrived from Dr. Hynes).</p><p> | ||
| + | In addition, the assay to test the specificity of the response of the transposon clones was repeated. In order to test the specificity of this response, an additional screen was performed using varying concentrations of hydrogen peroxide (to rule out activation by a general stress response in the cell) in addition to decanoic acid at a comparable concentration to that of the naphthenic acids used (to rule out activation due to sensing fatty acid compounds). The assay was carried out according to our methods documented in the <a href="https://2012.igem.org/Team:Calgary/Notebook/Protocols/tnscreen">protocol section</a>. This data was recieved after Wiki-Freeze had happened, and therefore will be analyzed at a later date. </p> | ||
| + | <h2>Week 23 (October 8-October 11)</h2><p> | ||
| + | Unfortunately, no transposon work was accomplished this week. | ||
| + | |||
| + | <h2>Week 24 (October 15-October 19)</h2><p> | ||
| + | Sequencing reactions came back as having no reads, so sequencing samples were resent. This happened multiple times this week, and it is unclear why reactions continue to fail. | ||
| + | |||
| + | <h2>Week 25 (October 22-October 26)</h2><p> | ||
| + | This week, data from the previously run assay was analyzed. The results of this can be seen below.</p> | ||
| + | <p align="justify"> | ||
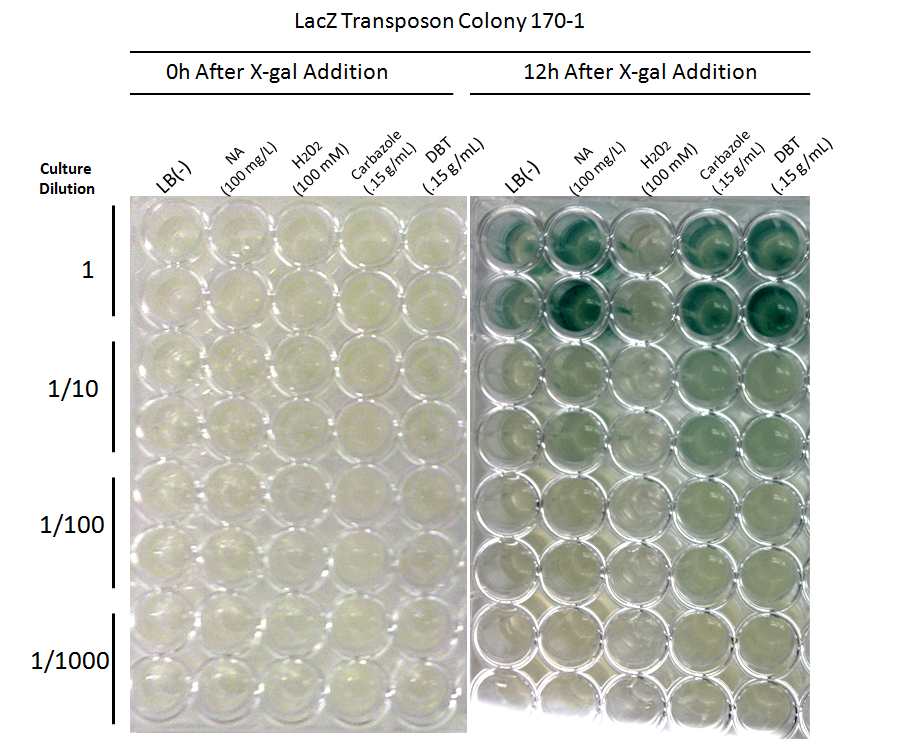
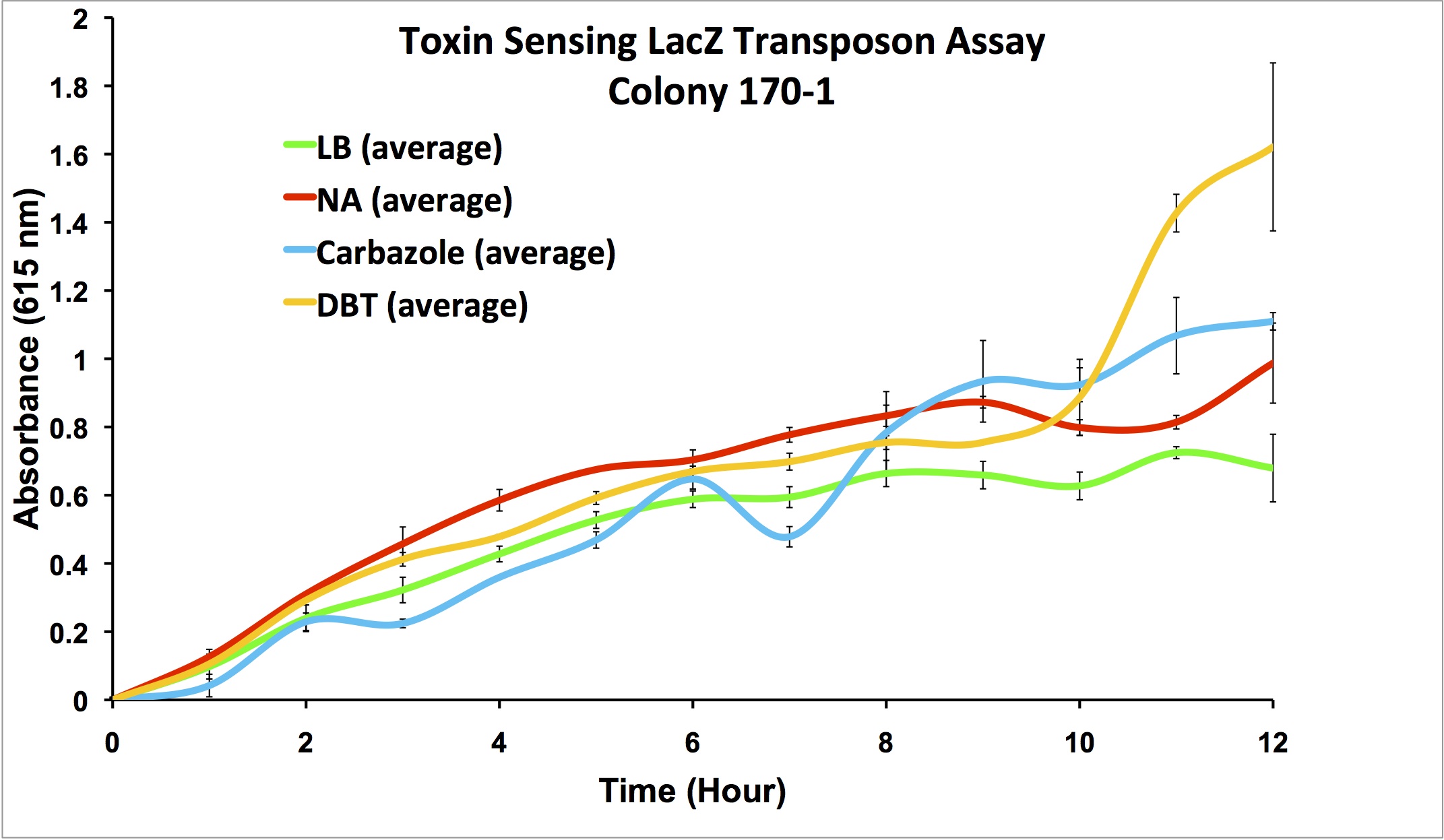
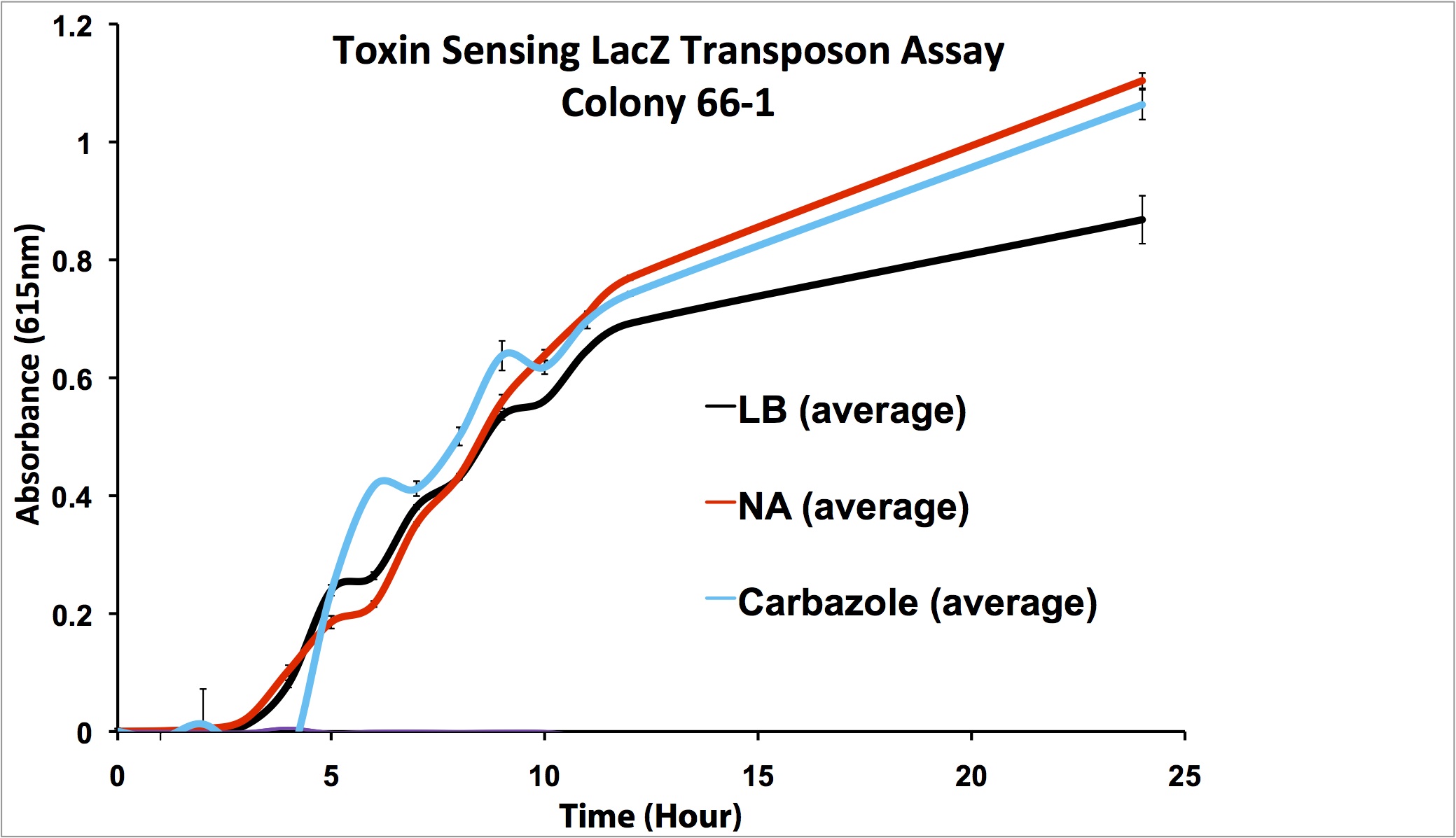
| + | </html>[[File:Ucalgary2012-FreddetectingTRANSPOSONstresstest.png|thumb|800px|center|Figure 8:Stress response screen on <i>P. fluorescens</i> Pf5 transposon mutants. Cells were inoculated in duplicate at different dilutions (shown are '''A:''' 66-1 undiluted, '''B:''' 66-1 at 1/10 dilution, '''C:''' 170-1 undiluted, '''D:''' 170-1 at 1/10 dilution) into LB as a control, LB containing varying concentrations of hydrogen peroxide, LB containing naphthenic acids at an environmental concentration, and LB containing decanoic acid at the same concentration as the naphthenic acids. 2 uL of 20mg/ml X-gal was added to the media and absorbance was read at 615nm (maximal absorbance of X-gal) every 4 hours for 12h. Higher absorbance was observed in the NA wells compared to the LB control, hydrogen peroxide, and decanoic acid for colony 66-1. Colony 170-1 showed a repressed response to naphthenic acids when compared to the LB control.]]<html><p> | ||
| + | These results show that colony 66-1 gives a response to naphthenic acids and other toxins that is not simply a response to fatty acids or a general stress response. Unfortunately, colony 170-1 does not show a useful reporter response.</p> | ||
| + | |||
| + | <p>In addition, electrochemical tests were conducted on the transposon clones this week. The results can be seen on the <a href="https://2012.igem.org/Team:Calgary/Project/Synergy"><b>Synergy</b></a> page. | ||
</html> | </html> | ||
}} | }} | ||
Latest revision as of 03:01, 27 October 2012


Hello! iGEM Calgary's wiki functions best with Javascript enabled, especially for mobile devices. We recommend that you enable Javascript on your device for the best wiki-viewing experience. Thanks!
Transposon Library Notebook
Week 1 (May 1-4)
This was the first week where we met with other team members and summarized the primary subprojects the team will be tackling this coming summer.
Week 2 (May 7-11)
During this week literature searches were performed.
Week 3 (May 14-18)
During this week literature searches were performed.
Week 4 (May 22-25)
During this week, strains of Pseudomonas fluorescens PF-5 were obtained. Two cultures were started by adding 500 µL stock to 10 mL LB media containing 50 mg/L ACROS Naphthenic Acids. These cultures were grown at 30°C overnight, shaking at 110 rpm.
Overnight cultures were then streaked on LB agar the following day with various types and concentrations of antibiotics in order to determine the susceptibility profile of the organism. This was necessary in order to determine what marker could be used on a transposon to allow for selection of organisms with sucessful transposon insertions. These plates were grown overnight to look for death or growth, and the following results were obtained;
| Gentamycin | Kanamycin | Chloramphenicol | Tetracycline |
| 25 µg/ml = no growth | 5 µg/ml = slight growth | 5 µg/ml = growth | 50 µg/ml = no growth |
| 50 µg/ml = no growth | 10 µg/ml = slight growth | 10 µg/ml = growth | 100 µg/ml = no growth |
| 100 µg/ml = no growth | 25 µg/ml = no growth | 25 µg/ml = growth | 200 µg/ml = no growth |
| 50 µg/ml = no growth | 50 µg/ml = growth |
Based on these results, it was determined that Kanamycin, Gentamycin, and Tetracycline could be used as the selectable marker on the transposon, while Chloramphenicol could not as the strain is naturally resistant. Glycerol stocks of the strains were also made at this time from a fresh overnight culture.
Week 5 (May 28-June 1)
At the beginning of this week we spoke to Dr. Michael Hynes, who was able to give us E. coli SM10 and SM17-1 cells containing the plasmid pOT182. This plasmid contains an E. coli origin of replication, allowing it to act as a suicide vector when transferred to a different bacterial species. pOT182 contains a Tn5 transposon element containing a promotorless lacZ gene, genes for tetracycline resistance as well as a beta-lactamase, transposase and an E. coli origin of replication. These elements are bordered by insertion element sequences which are recognised by the transposase. When transferred to a different host through conjugation, the plasmid itself can no longer replicate. The transposase however can recognise and transfer the sequence between the insertion elements in a cut-and-paste fashion randomly into the genome. In this fashion, the tetracycline and beta-lactam resistant traits would only persist in cells in which the transposon has jumped into the genome, allowing these antibiotics to select for transposon positive cells. The lacZ protein will only be produced if the transposon jumps in frame downstream of a promotor, and in this case would allow for a lacZ based assay of the promotors response. The E. coli origin of replication present in the transposon allows for the self-cloning of the transposon in a plasmid format after genomic digestion and circularization and transformation into E. coli, allowing for the sequence of bordering gene fragments to be determined easily and therefore mapping the transposon in the genome.
Cultures of E. coli SM10 and P. fluorescens PF-5 were grown up overnight in shakers at 37°C and 30°C respectively. SM10 was grown in LB + 10 µg/ml Tet, and PF-5 was grown in LB + 50 mg/L ACROS naphthenic acids. In the morning, the SM10 culture was subcultured (1/4) into LB without antibiotics and allowed to grow for an additional 4h. After this, 3 replicates of mating mixtures were made with 500 µl of each culture were mixed, and the cells were spun down and resuspended in 50 µl of LB. These samples were then plated in separate spots on LB agar. Additional spots were made in the same fashion with just PF-5 culture and just SM10 culture as controls, and the plate was incubated at 37°C overnight. In the morning, each spot was resuspended in 500µl sterile water + 25 µg/ml Tet, and dilutions of 1, 1/10, 1/100, and 1/1000 of each mating mixture was plated on Pseudomonas Isolation Agar (PIA) + 10µg/l Tet + 50 mg/L ACROS naphthenic acids. These cultures were allowed to grow at 30°C over the weekend.
The purpose of the PIA is to selectively allow the growth of the PF-5 strain, while killing off the donor SM10 strain. The tetracycline is designed to select for the positive transposon mutants in the PF-5 strain, as the only way that tetracycline resistance would be acquired (barring spontaneous mutation events) would be if the transposable element had jumped into the genome of the cell. We decided to use 10 µg/ml as the concentration of the tetracycline in the plates because we believed that 50 µg/ml would be too high for even strains carrying resistance to survive. Seeing as 10 µg/ml was effective for E. coli, we chose to try this. After streaking the PF-5 culture on the plate to test for its resistance however, it was found that the strain without the transposon was able to grow slightly on the plates, meaning that the concentration of antibiotic is not high enough to properly select for transposon mutants v.s. untransformed cells. In order to try to remedy this, the mating spots were resuspended in a mixture containing a higher dose of antibiotics. The results of this experiment are pending.
Week 6 (June 4-8)
When the plates from last week were examined, it was found that though the PIA was sucessful in inhibiting the growth of the E. coli donor strain, the original PF-5 strain was capable of growth on the media. Because of this, the plates were not selective towards cells containing the transposon insertion, and thus lawns of bacteria were seen on each of the plates.
Because of this, new selective media plates were prepared. These contained LB agar + 50 mg/L ACROS NA's + 50 µg each chloramphenicol and tetracycline. The chloramphenicol was used in order to kill off the SM10 donor strain, while the tetracycline was used at a concentration previously shown to kill off the PF-5 cells that did not contain a transposon insertion. The conjugation procedure previously described was repeated, with 1 replicate mating spot being plated in 4 different dilutions on the new selective media. These plates were grown overnight at 30°C. It was found that both SM10 and unmodified PF-5 were not capable of growth on the new selective plates, and colonies were found growing on all 4 of the dilutions for the mating spot.
Because the host and the untransformed cells were not capable of growth on the selective plates, it is believed that these colonies must represent sucessful transposition events, as this would be the only way that the tetracycline resistance would be transfered to the PF-5 cells (chloramphenicol would have no effect, as PF-5 cells are naturally resistant at this concentration, as previously shown). Because the cell density was too high, the 1 and 1/10 dilution plates were discarded, while the 1/100 and 1/1000 plates were stored at 4°C until the next step of the procedure, in which screening for naphthenic acid response will be performed.
Week 7 (June 11-15)
In order to test for a naphthenic acid response, a lacZ reporter system in the transposon will be utilized. Because the lacZ enzyme is capable of degrading X-gal, a dissacharide sugar containing lactose, into lactose, and an insoluble sugar (5,5'-dibromo-4,4'-dichloro-indigo), which appears blue. Therefore, in response to activation of a native gene promotor, in frame transposon insertions will produce lacZ at levels corresponding to the activation level of the gene, and these colonies will be able to utilize lactose as a carbon source as well as utilize Xgal as a substrate. Cells responding to naphthenic acids will therefore show blue pigmentation and be capable of growth on lactose. Those that do not respond should remain white, and perish when lactose is given as the only sugar source.
The concentration of naphthenic acids used in the test plates will be 4x less than the minimal inhibitory concentration (MIC). Therefore, a MIC assay for naphthenic acids with PF-5 cells was carried out overnight at 30°C at 2%, 1%, 0.5%, 0.25%, and 0.125% concentrations in LB media. It was found that PF-5 grew well at all these concentrations; therefore 1% was arbitrarily picked for the test conditions for naphthenic acid response plates.
Plates were made as follows, with Xgal spread on top:
| Wild-Type PF-5 Controls |
| M9 Minimal Media Alone, M9 Minimal Media + 0.4% Glucose, M9 Minimal Media + 0.4% Lactose, M9 Minimal Media + 1% NA's |
| Transposon Replica Plates |
| M9 Minimal Media + 0.4% Glucose + Chlor, M9 Minimal Media + 0.4% Glucose + Chlor + 1% NA's, M9 Minimal Media + 0.4% Lactose + Tet, M9 Minimal Media + 0.4% Lactose + Tet + 1% NA's |
Colonies from the 1/1000 transposon plate were replica stamped using velvet cloth onto the transposon test plates, and WT PF-5 was streaked onto the controls. The plates were allowed to grow over the weekend at 30 °C.
Week 8 (June 18-22)
The plates from the previous week were observed on Monday. WT PF-5 grew on M9 + Glucose, but not on any of the other controls, indicating that the wild-type strain could not utilize lactose, indicating that this could possibly be used as a selection measure for responses from the transposon mutants.
No growth was observed on any of the transposon test plates. This was believed to be likely from poor transfer during stamping, resulting from a poor contact with the plates in addition to a grease layer formed by the undissolved naphthenic acids. Because of this, a new method of creating plates had to be determined.
We tried to dissolve NAs in DMF and spread this mixture on top of agar plates as well as mix it into the liquid agar before pouring plates. Though this helped to solubilize the naphthenic acids, it was not as effective as increasing the pH of the media (pH 8 was found to be effective). We made M9 media at pH 8 before making the same set of test plates as before. It was found that the NAs stayed in solution when mixed, however came out of solution when the plates dried, leading to a grease layer on the plates as previously seen.
In another attempt to raise the pH, stock solutions of NAs dissolved in NaOH at pH 12 were made, and these were autoclaved alongside the media. When NAs were added to the NaOH, the solutions became cloudy and uniform, indicating they were in solution. When the NA stocks were added to the media before plate pouring, the NAs remained in solution even after the plates dried, indicating that this method would be sucessful in making test plates. Using this method, the following plates were created:
| Wild-Type PF-5 Controls |
| M9 Minimal Media Alone, M9 Minimal Media + 0.2% Glucose, M9 Minimal Media + 0.2% Lactose |
| Transposon Replica Plates |
| M9 Minimal Media + 0.2% Glucose + Chlor, M9 Minimal Media + 0.2% Glucose + Chlor + 0.05% NA's, M9 Minimal Media + 0.2% Glucose + Chlor + 1% NA's, M9 Minimal Media + 0.2% Lactose + Tet, 0.2% Lactose + Tet + 0.05% NA's, M9 Minimal Media + 0.2% Lactose + Tet + 1% NA's |
Xgal was spread on all the plates with NA's, however it was left out of the controls by mistake. Stamping was carried out as previously described from both the previously created 1/100 and 1/1000 dilution plates of transposon mutants (A new round of transposon mutagenesis was initiated, but the mating mixture was incubated at 30°C instead of 37°C, and no mutants were obtained on the selective plates). These plates were allowed to grow over the weekend at 30°C.
Week 9 (June 25-June 29)
Growth was observed from the selection plates incubated over the weekend at 30°C. Screening replica plates are made as before. The 1/1000 diluted transposon plate was used for the replica plating on all the screening conditions. The screening plates are incubated overnight at 30°C. The next day, no growth was observed on the negative control plates (M9 alone, M9 with 0.05% NAs), and no growth was observed on the positive control plates containing glucose. Similarly, no growth was observed on the M9+lactose (+Tet, with/without X-GAL, with/without 0.05% NAs). This suggests that either a large number of matings is needed before a desirable transconjugant is seen, or that the media condition requires modification. The plate surface is not as greasy as before, which suggests that the replica plating transfer process works. However, the pH of the media may affect the growth of transconjugants. Plans are made to modify the pH of the media before replica plating.
Week 10 (July 2-July 6)
Replica plating screening plates are made so that NA stock solutions (1% at pH 12 diluted to 0.05% final concentration) were added to the M9 agar solution. The pH is adjusted to 7.4 before autoclaving. The pH prior to adjusting is approximately 9.0. This may explain the poor growth observed in the previous trial. A new series of mating were started (2 mating spots), and incubated overnight at 37°C. The mating spots are scraped and serial diluted up to 1/1000, plated on LB+Chlor50+Tet50, and incubated overnight at 30°C. This time, the results are consistent with the previous attempt, where no growth was observed on the negative control plates (M9 alone, M9 with 0.05% NAs), and ample growth was observed on the positive control plates containing glucose. However, still no growth was observed on the M9+lactose (+Tet, with/without X-GAL, with/without 0.05% NAs)plates. This seems to confirm with previous hypothesis that a large number of matings are needed to screen the genome for NA sensitive elements. Alternatively, perhaps the NA concentration used here is too high for proper selection of the transconjugants that have a promoter element upstream of the transposon insertion. If the NA concentration used for the screen is too high, the promoter may be suppressed, and the lacZ is not expressed for cell survival. In fact, the lowest NA concentration used up to this point is 0.05% or 500 mg/L. The culturing conditions required to maintain Pseudomonas Pf-5's NA degrading abilities is LB+50mg/L. Therefore, to allow both cell survival and screen for the most robust and sensitive system, maybe the NA concentration should be lowered.
Week 11 (July 9-July 13)
The exact same experiment from the previous week has been repeated with two more mating mixtures (and their dilutions). However, we obtained the same results, with the positive and negative controls yielding predicted growth and no growth, respectively. But no growth was observed on the lactose plates. Furthermore, upon close examination of the selection plates after the conjugating/bipartite mating reaction (LB+Chlor+Tet), two colony morphologies can be seen: drier and larger colonies (resembling Pseudomonas), and smaller moist colonies (resembling E. coli). This suggests perhaps the LB+Chlor+Tet plates may be useful for selecting Pseudomonas transconjugants, but it does not prevent the donor E. coli to be efficiently killed. A final replica plating experiment is designed for next week, and Pseudomonas Isolation agar (PIA, just received)will be used for selection instead.
Week 12 (July 16-July 20)
Another mating reaction was set up, and plated on PIA+Tet50 to select for positive transconjugants. The colonies from the resulting mutants show only a single colony morphology consistent with Pseudomonas. From this, we know the selection is appropriate. The 1/1000 dilution of the mating reaction dilution is replica-plated on two sets of screening plates. The first set is prepared the same way as the previous week (NAs added and the media pH adjusted prior to autoclaving); the second set is the same except the NAs are added by spreading 100uL of a 50mg/L sterile stock NA solution in NaOH (pH12) prior to replica plating. The 1/1000 dilution plate of the mating reaction is replica plated on both plates (with the same controls as before). However, the same results are obtained as before, where no growth is observed on the M9+0.2% lactose+X-GAL or the M9+0.2% lactose+X-GAL+50mg/L NAs plates. Lastly, we had an interesting observation where overnight growth of the lactose screening plates showed no growth, if they are left for up to 48hr, some colonies can be observed; however, none of the colonies are blue. The problem may be that the M9 media conditions are not suitable for selecting the Tn mutants, or that the replica plating approach we used cannot effectively transfer all colonies. At this point, since no NA-sensitive strains are isolated, a new approach needs to be taken in order to find a tranposon insertion mutant that has an NA-sensitive promoter upstream. Since all possible approaches have been taken using the replica plating strategy, another mass screening method is needed.
Week 13 (July 23-July 27)
Even though, the previous screening method is quick and convenient, the waiting period is too long, and no real time data in terms of growth or lactose utilization can be observed/discerned. Therefore, a 96-well mass screening method is devised, where the conjugation mating reaction, and the selection on PIA+Tet50 are the same, but the screening step is different. The screening media is a M9 minimal media with 50 µg/mL tetracycline to maintain the genomic Tn insertions, 0.2% lactose to select for lactose utilization due to Tn insertions, and 50 mg/L NAs to maintain the strain's ability to use/degrade NAs (in short the media is M9+0.2%lactose+50µg/mL Tet+50mg/L NAs). On each 96-well plate, 94 colonies are inoculated and screened; the other two wells are used for negative and positive controls. The negative control well has the same media (M9+0.2%lactose+50µg/mL Tet+50mg/L NAs), with no colonies inoculated. The positive control has M9+0.2%glucose+50µg/mL Tet+50mg/L NAs, and is inoculated with a random colony.
Using this new method, any inoculated colony that grows in the liquid media can utilize lactose, and this should be because of a transposon insertion downstream of a promoter. To select for a NA sensitive promoter, each colony is inoculated into two plates with the same media components (as described above), except one has NAs, and the other does not. If a promoter is NA-sensitive, then it should not grow if NAs are absent from the media, as the NA mixture would activate the promoter. When NAs are present however, the NA-sensitive promoter would turn on, allowing it to utilize lactose, survive, and grow. As a first run of this method, a plate is set up using mutant colonies from a previous mating reaction. This plate contained M9+0.2%lactose+50µg/mL Tet+50mg/L NAs. The appropriate control plate with no NAs was not included as this is only to test feasibility of this approach. The plate is incubated over the weekend for 60hrs in a plate reader at 30°C.
Week 14 (July 30-August 3)
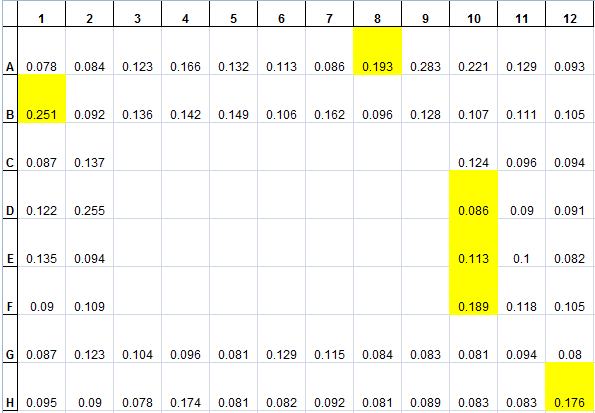
The data from the plate incubated from last week was collected. First, since the lid of the 96-well plate was kept on the plate to prevent dehydration of the media, there was significant condensation on the middle of the lid (covering about 30 wells), which prevented proper absorbance measurements of those wells (data from these wells were omitted). However, six wells/colonies demonstrated growth from a baseline. The following table is a display of the 96-well plate at the end point, with the colonies/wells that grow were highlighted.
The cultures from these six wells are washed with M9+0.2%lactose+50µg/mL Tet (no NAs) three times, and resuspended in 1 mL of M9+0.2%lactose+50µg/mL Tet. 500µl of each colony was inoculated into 5mL cultures with M9+0.2%lactose+50µg/mL Tet or M9+0.2%lactose+50µg/mL Tet+50mg/L NAs. These cultures are grown over 48 hours to observe growth (any strain that grows only when NAs are added would be noted). Also, the colonies are restreaked on PIA+Tet50 plates to ensure that the absorbance increase actually indicated growth (since the increase was so small at around +0.200). The results were not promising, on the restreaked plates, only 2 colonies (from H12 and B1) showed growth, this suggests some of the measurements were not very accurate. Also, in the culturing experiment, only 1 set of cultures (from 1 colony, H12) showed growth, but in both media conditions, which is not desirable.
Even though, no colonies were shown as NA sensitive, this result is still very promising, as this experiment shows that some colonies from the transposon mutagenesis can actually utilize lacZ, which demonstrates that the transposable element approach is an appropriate and feasible approach for screen promoters sensitive to environmental stimuli. (Please note, that mating reactions and mutant selection are conducted regularly every week, usually twice per week, with two mating spots each time, to provide mutants for these screens.)
Week 15 (August 6-August 10)
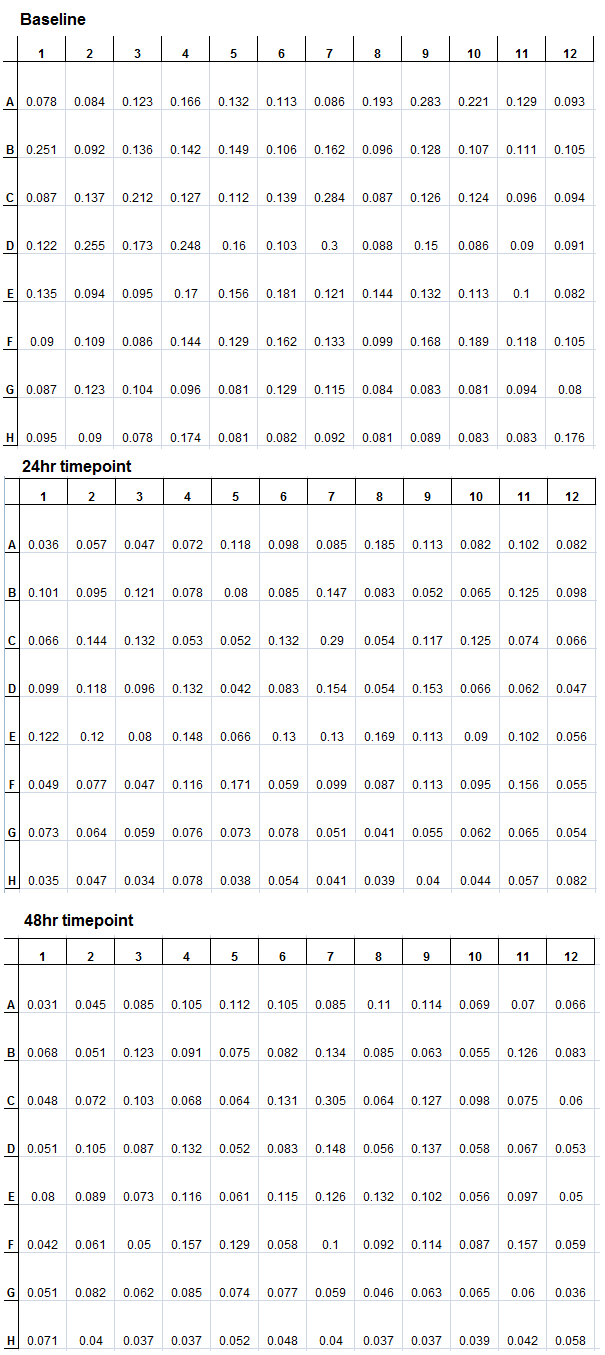
The same experiment from the previous week was repeated. However, the plate was not incubated in the plate reader, but incubated in a 30°C shaker, at 140rpm. At the baseline (beginning of incubation), 24hr, and 48hr, timepoint measurements were made. The results are as follows.
However, these measurements were made with the lid of the plate on. Even though there was no clear trend in terms of the growth in the absorbance data, it is possible to observe some wells become cloudy by eye. For instance, well F9 was cloudy at the end of the 48hr incubation, but the data showed no change in absorbance. The lid should be taken off in the future when making timepoint measurements. Also, the absorbance data can support the visual data.
Also, this week, two runs of the screening experiment was conducted with two plates run in parallel, one with NAs and the other without (the proper screening setup). However, it was found that the lactose solution (which was filter sterilized) used to make the M9 media for the screen was contaminated with yeast. Therefore, new media solutions and lactose solutions needed to be made. These data are not used because of this.
Week 16 (August 13-August 17)
New lactose and glucose stock solutions as well as new M9 screening media were prepared. The experiments from last week were repeated using proper solutions. Also, to prevent fogging of the plate lid and cross-contamination, all media solutions and plates were pre-warmed in a 30°C incubator before dispensing the media into the plates and colony inoculation. The new trial, again, has two plates, with the same wells (e.g. A1) inoculated with the same colony, where one plate has M9 screening media (with Tet and lactose) alone and the other plate containing 50mg/L NAs. Two runs were completed this week. The data from one of those trials are shown below.
In this particular run, only two colonies showed growth (D8 and H11), these were subcultured in 5mL of M9 screening media with NAs. An interesting observation is that there is more growth in the lactose+NAs plate than the lactose alone plate. This suggests that the NAs may play a role in activating the lacZ gene, and improving the survival rates. These results are consistently observed in the other trials conducted.
At this point, since time is limited, the entire mutant selection process needs to be more efficient. Therefore, the mating spots are diluted, and plated on a more selective media, containing PIA+50ug/mL tetracycline+50mg/L NAs+20 μg/mL X-Gal. This way, the NA-responsive and/or lactose-utilizing Pseudomonas transconjugants can be selected as they would appear blue on the media. This reduces the number of colonies to be inoculated, and improves the efficiency of the whole process. (Note: Wildtype Pseudomonas fluorescens pf-5 are always plated as positive control to observe the natural forward mutation rates, and to ensure that the amount of spontaneous tetracycline resistant Pf-5 mutants are at a reasonable level.
At this point, a large number of matings should be conducted and mutants selected, in order to provide enough of lactose-utilizing and NA-sensitive Tn-insertion mutants, and to cover as much of the Pseudomonas genome as possible
Week 17 (August 20-August 24)
No work on the transposon library was done this week, other than planning for the upcoming screen.
Week 18 (August 27-August 31)
This week, a large screen was accomplished. First, cultures of P. fluorescens Pf-5 and E. coli SM10 were grown up overnight at 30°C and 37°C respectively to a high optical density. The following morning, the E. coli was subcultured in a 1/4 dilution to dilute out the tetracycline, and grown for approximately 2 more hours. After this, 100 µL of each culture were mixed, spun down, and resuspended in 20 µL of media before being spotted onto LB plates.
In total, 500 separate mating spots were plated. These spots were allowed to grow at 37°C overnight. The following day, 2 mating spots were scraped up, combined, and resuspended in 500 µL PBS. 1/400 dilutions of these resuspensions were made, and 100 µL of these was plated onto selective plates consisting of PIA, 100mg/L NAs, and tetracycline. These plates had 40 µL of 20 mg/mL X-gal spread on their surface in order to allow for blue-white screening. 250 plates were made in total.
Week 19 (September 3- September 7)
Plates were left to grow for 2 days, after which blue colonies (24 in total) were selected and pinned in duplicate into 96-well plates for response testing. Initially, minimal media + lactose with and without NAs was used, however no growth in any of the wells was observed. Because of this, the screening protocol was altered such that LB with or without NAs was used instead of minimal media, and X-gal was used instead of lactose for screening- the idea being that if naphthenic acids were sensed, a blue color change would be observed relative to the negative LB control.
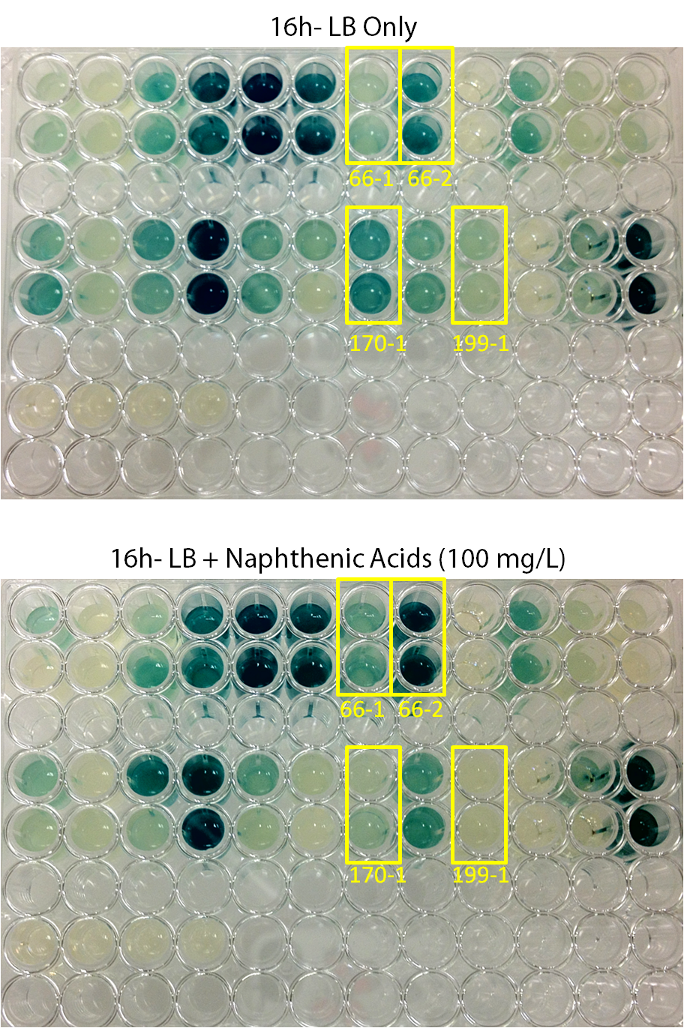
When results were observed it was found that 4 colonies showed clear differential regulation in response to naphthenic acids: 66-1, 66-2, 170-1, and 199-1. Therefore, these colonies will be used in further screening to test the specificity of the response.
Week 19 (September 10- September 14)
This week, further screens on the previously identified four hits were performed. These involved the use of different toxins at environmentally relevant concentrations to determine if the sensing response was specific to naphthenic acids, or if a sensory response to general toxins had been found. In addition, hydrogen peroxide was used in one of the media samples in order to attempt to rule out a general stress response by the cell.

Due to these results, further screens on these two colonies will be performed, using lower hydrogen peroxide concentrations to rule out a general stress response, and decanoic acid to rule out a response to fatty acids.
Week 20 (September 17- September 21)
Further screens were conducted, however due to cells drying out in the plate the results were invalidated. Because of this, the screen will be repeated, and explained in further detail when this is done.
Week 21 (September 24- September 28)
Genomes of 66-1 and 170-1 were isolated, digested with BglII and with XbaI, and religated before being transformed into E.coli. Cells were plated onto tetracycline plates to isolate cells containing a ligation product with the transposon present. These colonies have been miniprepped, and are awaiting being sent for sequencing to determine which genes the transposon has been inserted into.
Week 22 (October 1-October 5)
Minipreps from previous weeks of the transposon self-cloned plasmid were nanodropped, and it was determined that no DNA had been isolated. Therefore, colonies were re-prepped, and sent for sequencing (as sequencing primers have arrived from Dr. Hynes).
In addition, the assay to test the specificity of the response of the transposon clones was repeated. In order to test the specificity of this response, an additional screen was performed using varying concentrations of hydrogen peroxide (to rule out activation by a general stress response in the cell) in addition to decanoic acid at a comparable concentration to that of the naphthenic acids used (to rule out activation due to sensing fatty acid compounds). The assay was carried out according to our methods documented in the protocol section. This data was recieved after Wiki-Freeze had happened, and therefore will be analyzed at a later date.
Week 23 (October 8-October 11)
Unfortunately, no transposon work was accomplished this week.
Week 24 (October 15-October 19)
Sequencing reactions came back as having no reads, so sequencing samples were resent. This happened multiple times this week, and it is unclear why reactions continue to fail.
Week 25 (October 22-October 26)
This week, data from the previously run assay was analyzed. The results of this can be seen below.

These results show that colony 66-1 gives a response to naphthenic acids and other toxins that is not simply a response to fatty acids or a general stress response. Unfortunately, colony 170-1 does not show a useful reporter response.
In addition, electrochemical tests were conducted on the transposon clones this week. The results can be seen on the Synergy page.
 "
"