Team:Calgary/Notebook/Denitrogenation
From 2012.igem.org
Anyakornilo (Talk | contribs) |
|||
| (4 intermediate revisions not shown) | |||
| Line 102: | Line 102: | ||
<p>Constructs that we continued to work on this week included <i>AmdA</i> with <a href="http://partsregistry.org/Part:BBa_J13002">J13002</a>, <i>carAa</i> with <a href="http://partsregistry.org/Part:BBa_J13002">J13002</a>, and <i>carAc</i> with <a href="http://partsregistry.org/Part:BBa_B0034">B0034</a>. Some sequencing was sent, but no positive results were obtained. A new approach to biobrick the <i>ant</i> operon was implemented this week. A new forward primer was designed that would anneal about 1500 base pairs upstream of the <i>ant</i> operon, avoiding the region of homology that is repeated at various points on the plasmid. Once this region is successfully amplified we could use this amplified DNA as a template for another PCR reaction which would use the normal <i>ant</i> primers. Since the template would no longer include regions of homology these primers should be free to work properly and amplify the <i>ant</i> operon. The PCR using the new primers was started this week, and although amplification at 4500 base pairs was observed as expected, there was also a lot of non-specific amplification. This means that to purify the amplified DNA that we want and avoid the other products a dreaded gel extraction had to be performed which, as usual, did not work. At this point it was decided that further attempts to amplify the <i>ant</i> operon would be futile and our efforts would be better served working on constructing the parts we do have. Unfortunately no progress was made in these weeks on construction. We have also obtained GC-MS readings from the carbazole degradation experiment that showed that <i>Pseudomonas</i> LD2 can indeed degrade carbazole, however only under low glucose conditions.</p> | <p>Constructs that we continued to work on this week included <i>AmdA</i> with <a href="http://partsregistry.org/Part:BBa_J13002">J13002</a>, <i>carAa</i> with <a href="http://partsregistry.org/Part:BBa_J13002">J13002</a>, and <i>carAc</i> with <a href="http://partsregistry.org/Part:BBa_B0034">B0034</a>. Some sequencing was sent, but no positive results were obtained. A new approach to biobrick the <i>ant</i> operon was implemented this week. A new forward primer was designed that would anneal about 1500 base pairs upstream of the <i>ant</i> operon, avoiding the region of homology that is repeated at various points on the plasmid. Once this region is successfully amplified we could use this amplified DNA as a template for another PCR reaction which would use the normal <i>ant</i> primers. Since the template would no longer include regions of homology these primers should be free to work properly and amplify the <i>ant</i> operon. The PCR using the new primers was started this week, and although amplification at 4500 base pairs was observed as expected, there was also a lot of non-specific amplification. This means that to purify the amplified DNA that we want and avoid the other products a dreaded gel extraction had to be performed which, as usual, did not work. At this point it was decided that further attempts to amplify the <i>ant</i> operon would be futile and our efforts would be better served working on constructing the parts we do have. Unfortunately no progress was made in these weeks on construction. We have also obtained GC-MS readings from the carbazole degradation experiment that showed that <i>Pseudomonas</i> LD2 can indeed degrade carbazole, however only under low glucose conditions.</p> | ||
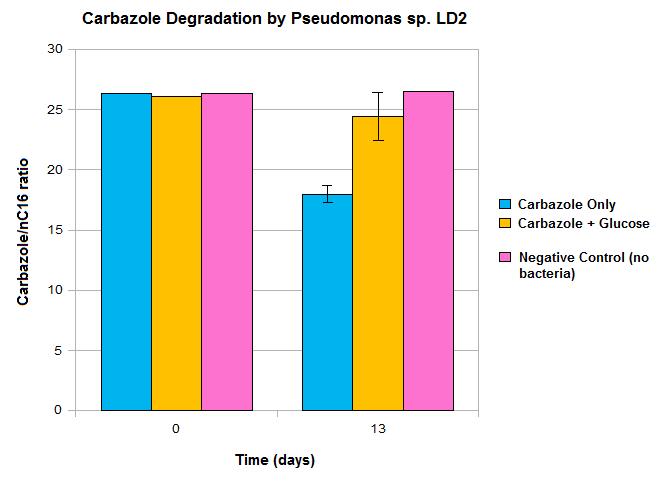
| - | </html>[[File:LD2 degradation september data calgary12.jpg|500px|thumb|Carbazole loss over 13 days when cultured with <i>Pseudomonas sp. LD2</i> with and without the addition of glucose.|center]] | + | </html>[[File:LD2 degradation september data calgary12.jpg|500px|thumb|Carbazole loss over 13 days when cultured with <i>Pseudomonas sp. LD2</i> with and without the addition of glucose.|center]]<html> |
| + | <h2>Weeks 23-26 (October 1-October 24)</h2> | ||
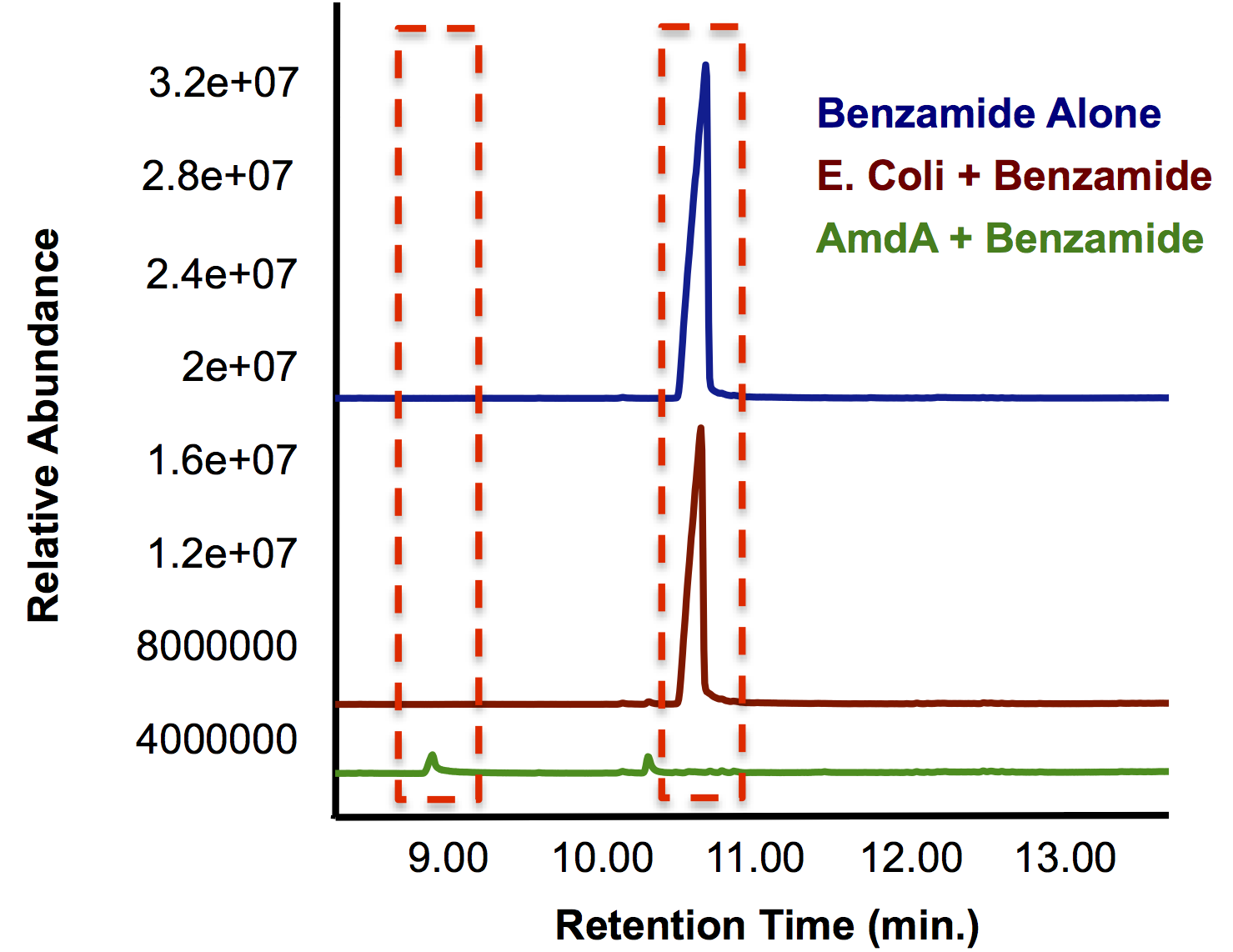
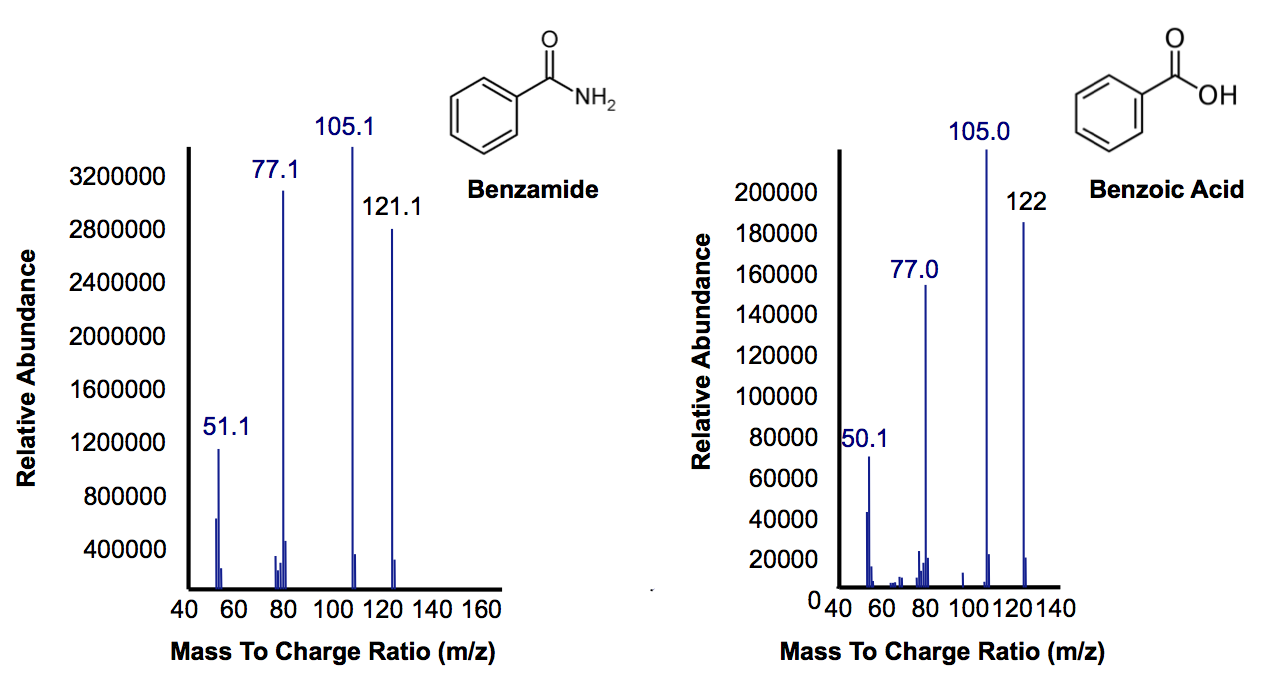
| + | <p>Most of this time was spent attempting the same construction of <i>AmdA</i> with a TetR promoter as described above. After <a href="http://partsregistry.org/Part:BBa_K902041">this construct</a> was finished, we designed two experiments to characterize this biobrick. One was an in vivo experiment in which <i>E. coli</i> cells containing the biobrick were assayed with 3 compounds containing a primary amine or amide. Benzamide, which has similar structure to the naphthenic acids being targeted by other parts of the hydrocarbon project, and butyramide were previously shown to be good substrates for the AmdA enzyme (Hirrlinger et al, 1996). Benzamidine was also tested to explore the array of substrates that could possibly be degraded by the enzyme. The other approach we took was to create a cell lysate from <i>E. coli</i> that was grown up and resuspended it in a lysis buffer. This was assayed with the same 3 compounds to guard against the possibility that the in vivo system lacked the appropriate transfer proteins. Hopefully this stuff works.The data below from the GC shows that one compound gets completely degraded by the cells containing the <i>AmdA</i> gene as well as a new product being formed in this group. The MS data shows that the compound being obliterated by amidase is in fact benzamide and that the new product being formed is benzoic acid. Benzoic acid results from replacing the NH<sub>2</sub> with an OH group, which is the expected function of the AmdA enzyme. The protocol for this experiment can be found <a href="https://2012.igem.org/Team:Calgary/Notebook/Protocols/AmidaseAssay">here</a> </p> | ||
| + | </html>[[File:Calgary AmdA Chromatograph.png|thumb|420px|centre|GC chromatograph demonstrating the degradation of benzamide by <i>AmdA</i> compared to an <i>E. coli</i> control. Additionally, a smaller peak with an earlier retention time is produced solely in the sample run.]] | ||
| + | [[File:Calgary AmdA MSData.png|thumb|420px|centre|Mass Spectra for the peak at a retention time of 10.5 minutes from Figure 7 representing the substrate benzamine. Additionally, the peak at retention time of 9 minutes represents benzoic acid based on its mass suggesting that their is a small accumulation of product.]]<html> | ||
| + | |||
| + | |||
| + | </html> | ||
}} | }} | ||
Latest revision as of 00:16, 27 October 2012


Hello! iGEM Calgary's wiki functions best with Javascript enabled, especially for mobile devices. We recommend that you enable Javascript on your device for the best wiki-viewing experience. Thanks!
Denitrogenation Journal
Week 1 (May 1-4)
The following section covers wetlab aspect of our overall project focusing on microbial conversion of naphthenic acids into economically-valuable hydrocarbons. The approach taken in this endeavour will be from four strategic starting points - ring cleavage, decarboxylation, denitrogenation, and desulfurization. Overall, the 'hydrocarbons' aspect of the project is a critical one to our overall design and construction of a bioreactor capable of not only detecting but also converting naphthenic acids and other toxins to useful or at least innocuous compounds.
Week 2-3 (May 7-18)
In the first two weeks of iGEM our group has focused on reviewing literature regarding the bioremediation of nitrogen groups attached to naphthenic acids. The most prevalent N heterocycle is carbazole, representing 75% of total nitrogen by mass. The upper pathway of carbazole biodegradation is catalyzed by the enzymes coded for by the car operon, CarA (CarAaAcAd), CarB (CarBaBb), and CarC. These enzymes convert carbazole to anthralinic acid. The lower pathway is catalyzed by the enzymes of the ant operon, antA, B, and C, yielding catechol while releasing CO2 and NH3. The car and ant operons are both regulated by the Pant promoter which is induced by the protein, antR. carAa also has its own promoter which is not induced by antR (Diaz & Garcia, 2010). We have also investigated an alternative pathway using CarA combined with an amidase (amdA) that selectively cleaves NH2 from an intermediate of the car pathway (Xu et al, 2006). This could bypass much of the car/ant pathway and is possibly more efficient.
We have decided to use Pseudomonas resinovorans and Rhodococcus erythropolis to derive these genes from. carABC and antABC from P. resinovorans has been shown to have a wide range of nitrogen containing substrate specificity. R. erythropolis contains the amdA gene that we wish to use, and some evidence suggests that it may also be able to degrade sulfur rings through its carABC pathway.
Week 4 (May 22-25)
This week we reviewed the primers listed in the database and also designed some new ones. Primers for carAa, carAc, and carAd were designed individually, while primers for CarBaBbC and antABC were designed to encompass multiple genes in a sequence. These primers were designed to be used on Pseudomonas putida which we decided to use as our gene source since it was available to us as opposed to ordering Pseudomonas resinovorans. We also designed a primer for the AmdA gene from Rhodococcus erthyroplois. In addition to ordering these primers we also ordered the nitrogen containing compounds that we will need to test these enzymes on. We decided on using carbazole to make sure the enzymes can perform their natural function as well as pyrrolidine to test them on a similar ring structure. We also ordered cyclohexamine in order to independently test the function of the alternative AmdA pathway. Finally, we decided to eventually order 4-Piperidine butyric acid hydrochloride to test how the enzymes will work on nitrogen containing naphthenic acids. However, we decided since it is a very expensive compound we would wait to make sure the enzyme's work on more simple compounds before ordering it.
We also started our work in the wet lab by plating colonies and making an overnight culture of -80 freezer stock Psuedomonas putida on Thursday. We also resuspended the primers for carAa, carAc, carAd, antA, antB, and ant C that were already in the database. We were able to use the colonies that grew on the streak plates to start a colony PCR to attempt to isolate each of these genes on Friday.
Week 5 (May 28 - June 1)
On Monday and Tuesday we used the database primers to attempt to isolate carAa, carAc, carAd, antA, antB, and antC from the Psuedomonas putida we plated last week. carAd, antB, and antC were all put in the gradient PCR machine to account for the wide range of their primers’ melting points, while the others were done via regular PCR. Unfortunately, no positive results were obtained from these reactions. The PCR on carAc and antA was repeated on Wednesday, still not giving positive results. Finally, we attempted to use a salt concentration gradient on the PCR reaction for carAc and antA, using concentrations that ranged from 1.0 microlitres/tube to 2.0 microlitres/tube in increments of 0.2 microlitres/tube. This helped as carAc showed bands in samples that had concentrations of 1.2 and 1.4 microlitres/tube. antA also showed bands, however they were not the correct size, indicating contamination and/or non-specific annealing of the primer. The positive control also showed bands of the correct size. Earlier in the week we also made overnight cultures of 3 environmental strains (28, 29, 30) of Psuedomonas putida from -80 glycerol stock. On Friday we performed a genomic prep on these cultures, and plan on attempting to amplify genes from the isolated DNA next week.
Week 6 (June 4 - June 8)
We started off this week by determining the DNA concentration of our genomic prep samples from last week using the nanodrop. DNA concentration for all three P. putida strains was at least 1000 ng/microlitre, well above what was needed for PCR. 1/2 and 1/3 dilutions were prepared for all three strains so as not to have an excess of template DNA in PCR reactions. PCR was performed on all 3 strains using primers for carAc, carAd, antA, antB, and antC using 6 replicates per gene. The only successful amplification appeared to be antB and carAc, both from strain 28 (with weaker bands in strain 29). We then performed another PCR, just on those two genes with an increased amount of Taq polymerase to hopefully get enough amplified DNA to move forward with. This resulted in strong bands for both at the expected size. We then performed PCR purification using the Qiaquick kit and obtained samples containing 33.5 ng/microlitre of antB DNA and 129 ng/microlitre of carAc DNA. These concentrations were both sufficient to begin a restriction digest and ligation of these parts into vector pSB1C3. Next week we hope to verify the results of the restriction digest, continue to amplify carAc and antB from strain 28, and hopefully submit a biobrick for sequencing.
Week 7 (June 11 - June 15)
The purified PCR products for carAc and antB were digested with restriction enzymes EcoRI+SpeI, EcoRI+PstI, and XbaI+PstI. However, only pSB1C3 plasmids that had been digested with EcoRI+SpeI and EcoRI+PstI were available to attempt ligation. Gel results were inconclusive on the restriction digest product as the plasmid size appeared much larger than expected. However, transformation was still attempted on the ligation products for both genes (both using EcoRI+SpeI ligation into pSB1C3 plasmid). The transformation products were plated onto chloroamphenicol (chlor) plates to select for colonies that had the chloro resistance gene on the pSB1C3 plasmid. Also this week another round of PCR was performed for the car and ant genes, however all, but carAc showed bands in the negative control lane, possibly indicating contamination or the formation of primer dimers. The carAc bands were fairly weak and a PCR purification resulted in very low DNA concentration, insufficient to move onto restriction digest. Next week's plan hinges heavily on the result of the transformation from this Friday.
Week 8 (June 18 - June 22)
The results of our transformation on carAc and antB from last week showed mostly red colonies indicating that the ligation was unsuccessful and that the PSB1C3 plasmid simply closed on itself with no insert. However, there were three colonies for carAc that were white (cut with both X+P and E+P). Colony PCR was performed on these three colonies using biobrick primer sets to verify that the part had been inserted in these colonies, but unfortunately all PCR results were negative indicating an unsuccessful ligation.
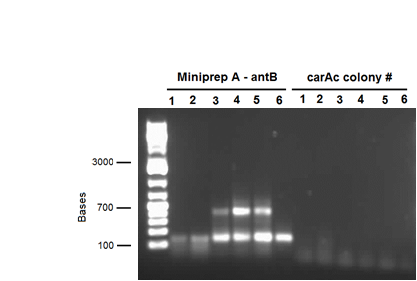
We also performed a miniprep on Pseudomonas putida strain #28 this week using a home-made protocol rather than the Qiagen kit. Miniprep A had a DNA concentration of 1539.8 ng/μL and Miniprep B had DNA concentration of 1001.2 ng/μL possibly indicating that there may have been a lot of genomic DNA contamination. However miniprep A product was still used as a PCR template for a reaction attempting to isolate carAc, carAd, antA, antB, and antC. Special conditions for this PCR included replacing 60 μL of water with 60 μL of betaine heated to 37 degrees Celsius and running a Mg gradient ranging from 0.5 μL - 2.5 μL per tube for each gene. Of these only antB had bands of the right size in 3 of the lanes (Mg concentrations of 1.5, 2, and 2.5 μL). However, all of the antB bands (including the negative control) contained a contamination band at around 100 bases, probably due to the formation of primer dimers. This forced us to do a gel extraction rather than a PCR purification. This gel extraction did not work as the nanodrop results indicated high amounts of agarose contamination. Another PCR was performed using the same conditions, but using miniprep B instead as there was no more miniprep A product remaining. Unfortunately there were no bands for antB, only the 100 base pair contamination bands.
Week 9 (June 25 - June 29)
This week we modified our PCR protocol to adjust for the homemade Taq polymerase we were using. Template DNA was boiled for 10 minutes on its own to replace the initialization step. This is because this polymerase could not tolerate the 95 degree heat during the regular initialization step. Another modification we made was to boil our primers for 5 minutes prior to adding them to the master mix. This was done to discourage the formation of primer-dimers. Finally we also attempted a touch-down PCR which started at an annealing temperature greater than the Tm of our primers and gradually worked its way down to below the Tm of the primers. This was done to increase the specificity of our reaction and also eliminate the formation of primer-dimers. Unfortunately these techniques still saw the formation of dimers, preventing us from performing a PCR purification. Again, the gel purification procedure did not lead to successful purification of our target gene. Due to the problems we have had so far our goal for next week is to perform an assay experiment to try and determine which of the Pseudomonas putida strains we have access to can actually degrade carbazole and therefore contain the pCAR1 plasmid.
Week 10 (July 2-July 6)
The goal for this week was to grow different strains of Pseudomonas in media containing B-N media + glucose with only carbazole as a source of nitrogen. In theory only those strains which contained the pCAR1 plasmid would be able to use carbazole as a nitrogen source and thrive in this media. The strains we tested from glycerol stock included P. putida LD1, P.putida environmental strains 28-30, and P. fluorescens PF5. Each of these strains had two cultures grown; one with carbazole and one without it. E. coli grown with carbazole was used as a negative control as it does not contain the pCAR plasmid. At timepoints of 22, 46, and 117 hours of growth samples were taken to do an OD600 reading to measure bacterial growth and had an ammonia assay performed on it. The detection of ammonia acted as a proxy for detecting carbazole degradation as it is an end-product of the pathway. With this in mind, an additional control strain of LD1 was grown in media containing glucose and ammonia. This is to detect the possibility that Pseudomonas can use ammonia as a source of nitrogen after it has degraded carbazole which would be problematic as far as using ammonia levels to determine carbazole degradation. So far we have been having difficulties reading the results of the ammonia assay with a plate reader and may resort to testing our samples using a different assay protocol next week. The OD600 readings were initially very high at 22 hours and then dropped at 46 hours.
Week 11 (July 9-July 13)
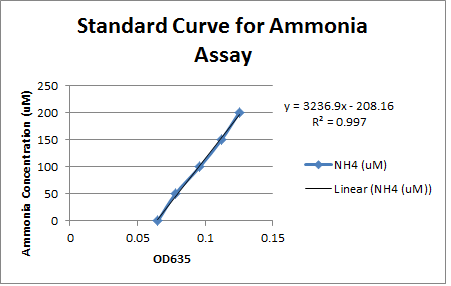
This week we took our last readings from the experiment that we started last week. The OD600 readings taken at 117 hours showed minimal growth over the weekend for all strains except for environmental strain #29 (with carbazole) which increased from 0.086 to 0.313. No other strain showed an increase of 0.100 or greater. This shows that only strain #29 was able to thrive in the carbazole media, possibly indicating that it contains the pCAR plasmid enabling it to use carbazole as a nitrogen source. To verify this we used a new ammonia assay protocol to measure the production of ammonia in the different cultures and theoretically how much carbazole was degraded. Unlike the previous assay we were able to produce a standard curve with a consistent slope based on samples with known amounts of ammonia. However, the only samples that showed any elevation of ammonia levels above the blank were the LD1 samples that were cultured with ammonia chloride to control for the possibility of pseudomonas using ammonia as a nitrogen source. The ammonia levels in this sample were relatively constant which would indicate that the LD1 was not using ammonia as a nitrogen source. Unfortunately the fact that none of the test samples had elevated ammonia levels did not give us any evidence that these strains contain the pCAR plasmid.
Week 12 (July 16 -July 20)
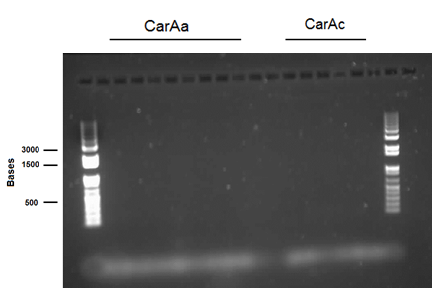
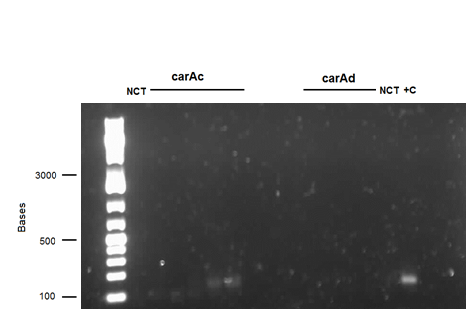
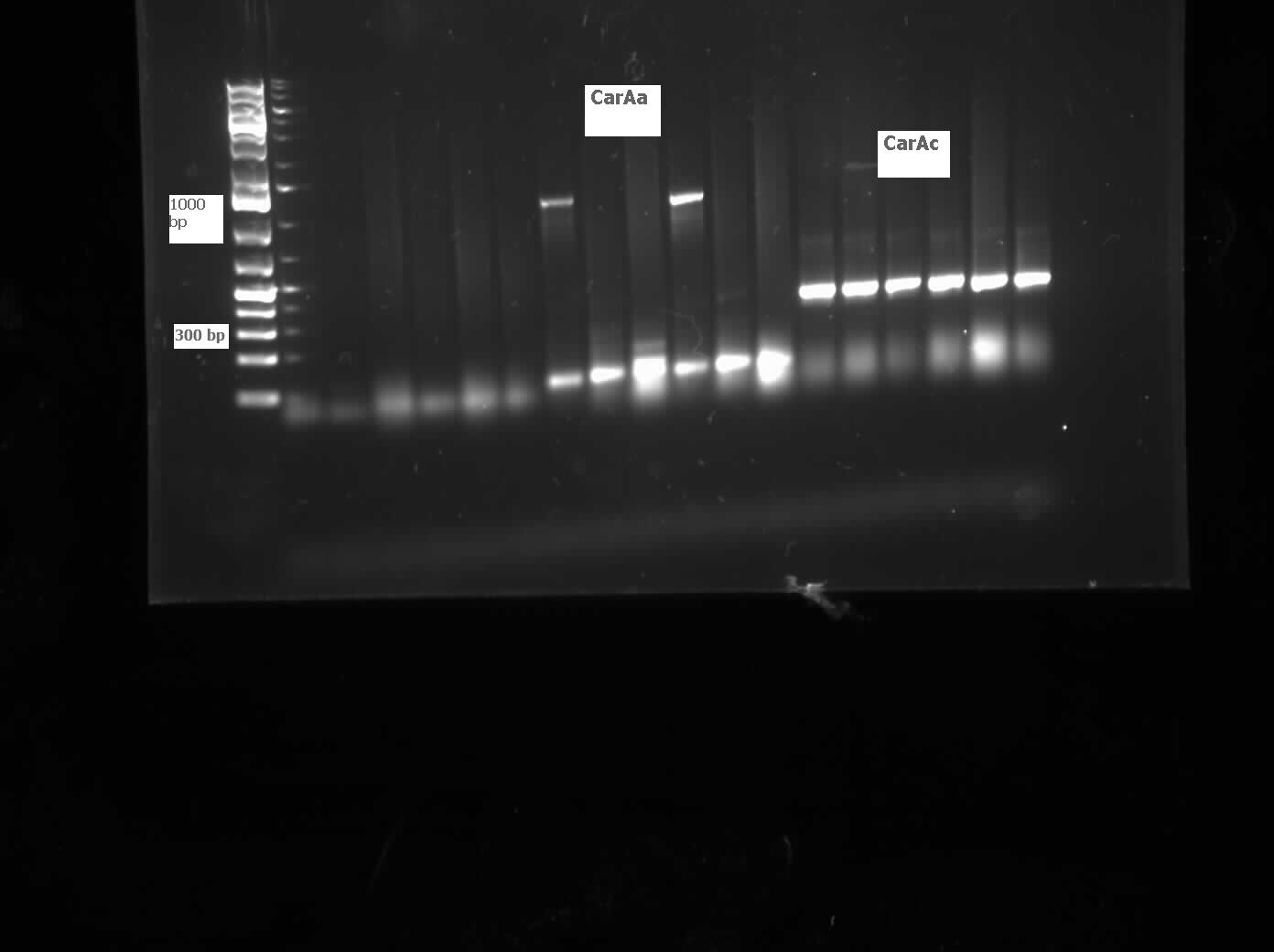
We received a new strain of Pseudomonas LD2, this week that had previously been used to study the pCAR1 plasmid. It was grown up in PCA media and had its colonies used for colony PCR. We also received our new primers this week, further increasing the chances of successful PCR. Using the new strain and the new primers carAa, carAc, and carAd all showed at least some bands of close to the correct size. After PCR purification we obtained DNA concentrations of 21.3 ng/μL for carAa, 41.9 ng/ μL for carAc, and 32.8 ng/μL for carAd. These products were then restriction digested using EcoRI + PstI sites.
Week 13 (July 23 - July 27)
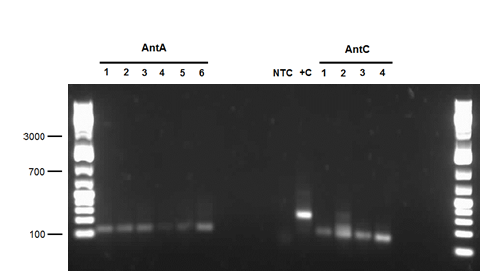
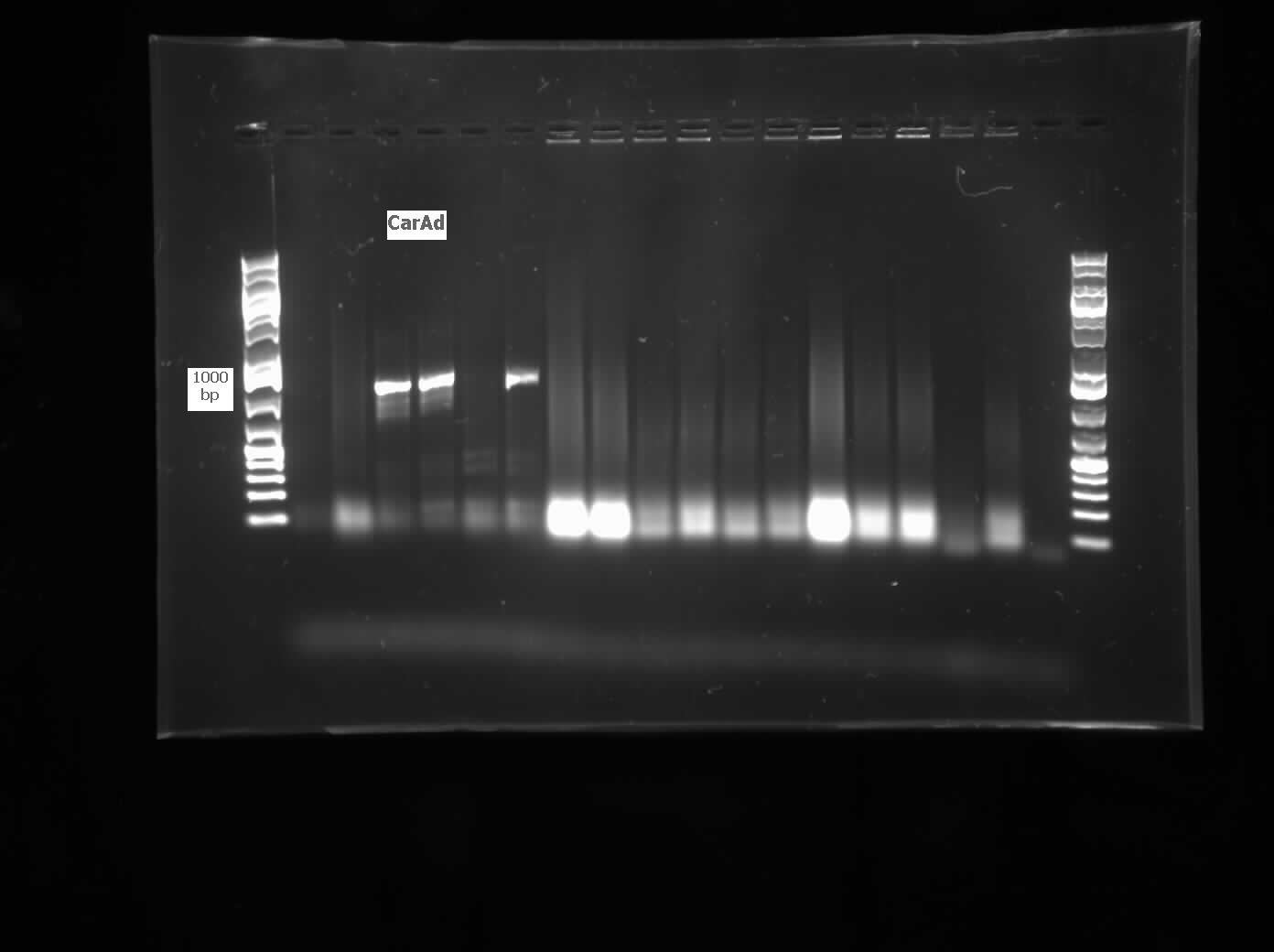
The restriction digested car genes were ligated into vector pSB1C3 and transformed into competent E. coli cells. White colonies grew for carAa and carAc suggesting that ligation and transformation were successful, however no colonies grew for carAd suggesting that the transformation did not work. Colony PCR was performed to confirm the presence of these genes within the vectors, however only carAc was positive. The colonies containing the carAc plasmid were then mini prepped and restriction digested to isolate the plasmid and send for sequencing. Further attempts at transforming carAa and carAd were not successful. Another goal was to perform colony PCR on the P. putida to isolate the antABC genes using both the primers for the entire operon and those for the individual genes. No attempt showed any bands, although salt and temperature gradients were used as well as different types of polymerase. LD2 cultures were grown up to be plasmid prepped and genomic prepped in order to have a higher concentration of template DNA in the PCR master mix and more PCR will be attempted on these genes next week. Mutagenesis primers were also designed this week to remove the illegal restriction cut sites in the carAd and antB genes (NotI and 2xEcoRI respectively). Only 1 mutagenesis primer was designed for antB as one of its EcoRI sites was very close to the beginning of the gene and could be removed using a new primer. During this process it was also discovered that the original ant primers were designed for the wrong genes and therefore do not match the intended sequence. Although the new antABC primers were designed using the correct gene the PCR may not be working due to the length of the operon so the design of new individual primers may be necessary. Finally, we also aimed to grow up the freeze dried culture of Rhodococcus erythropolis (source of the AmdA gene) that we received from DSMZ. The reactivation media was produced and autoclaved to begin the process next week.
Week 14 (July 30 - August 3)
We received the sequencing results this week for carAc which came back as a 100% match to the expected nucleotide sequence. We also successfully ligated both carAa and carAd into pSB1C3 and transformed into top 10 E. coli cells. Colony PCR verified the presence of the biobrick plasmid in these colonies. These were both miniprepped, digested and sent for sequencing by the end of the week. Using a salt gradient, addition of DMSO, and the KAPA buffer specifically designed for GC-rich amplicons we were able to amplify the ant operon from a genomic prep of LD2. However, all attempts at ligating and transforming this sequence were met with the unfortunate presence of red colonies (indicating failed insertion into the plasmid). Next week we will recut a pSB1C3 vector with Xba and Pst to attempt to mitigate this problem. We also reactivated the R. erythropolis following the DSMZ protocol for reviving freeze-dried cultures that can be found here using DSMZ media #1 and grew it on plates made from DSMZ media #220 at 28 degrees. Colonies grew very quickly and were used for colony PCR and to make overnight cultures for downstream applications. The colony PCR using the amidase primers was unsuccessful, so a genomic prep was performed on the overnight culture. Unfortunately we lacked some of the required equipment and the genomic prep did not work, but with a few procedural modifications and the right equipment we are confident it will work next week. A glycerol stock of the R. erythropoplis was also made from another overnight culture. We also started construction on the carAc part this week by inserting a ribosome binding site (B0034) in front of the gene. Finally, since the ant operon was successfully amplified by PCR we designed mutagenesis primers for the 2nd EcoRI site in the operon rather than making new individual primers. We also designed a mutagenesis primer to remove the SpeI site in AmdA. Now that we have conclusively proven that the LD2 strain contains the pCAR1 plasmid we aim to demonstrate its ability to degrade carbazole and other nitrogen containing heterocycles using an assay experiment similar to the one performed in Week 10.
Week 15 (August 7 - August 10)
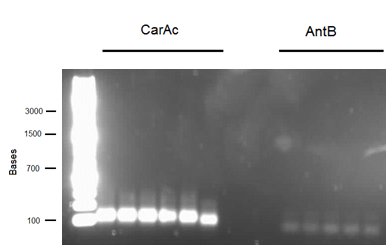
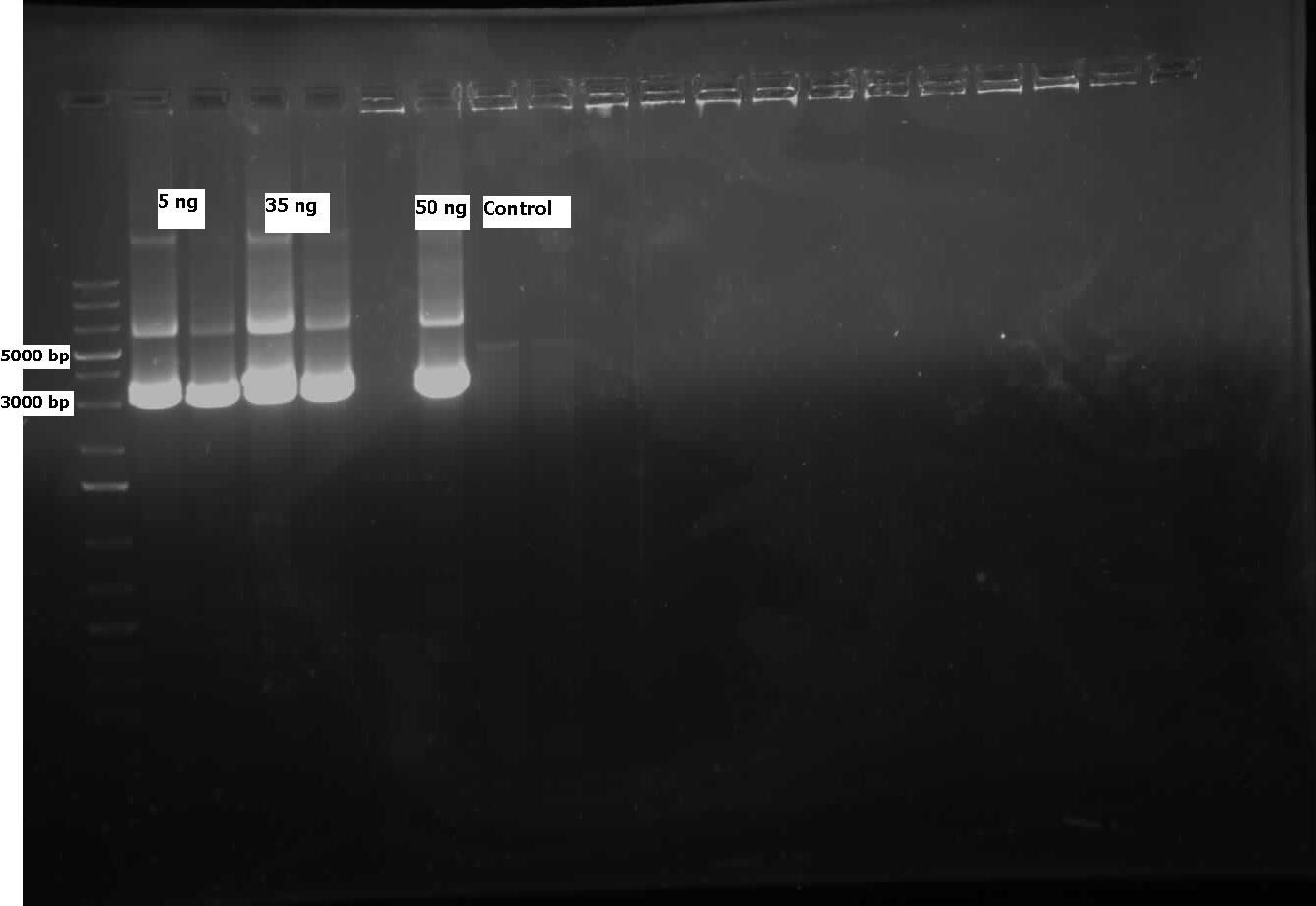
This week we received the sequencing results for carAa and carAd. carAd was a match, however carAa did not match at all and had misplaced biobrick restriction enzyme sites. Due to this we decided to restart the biobricking process for carAa all the way back to PCR from the plasmid. By the end of the week we had progressed to the point of transforming it into competent E. coli cells, however a thick lawn of bacteria had grown overnight making it impossible to pick colonies. A streak plate was made from this plate to attempt to get less dense growth. Since the sequencing came back positive for carAd the next step was to perform site directed mutagenesis on its NotI restriction enzyme site using the primers that were designed previously (for site-directed mutagenesis optimization, please refer to Desulfurization Journal Week 14). The PCR was successful, showing very bright bands at 3000 bp, and the PCR product was digested with DpnI to remove parental DNA. Transformation of this product will indicate whether the mutagenesis was successful. We were also able to successfully perform a genomic prep of Rhodococcus erythropolis and amplify the AmdA gene this week. It was ligated into pSB1C3 and transformed into competent E. coli by Friday, and a confirmation colony PCR was run on Friday evening to verify that the gene was inserted. These results will be available by Monday. After many attempts the ant operon was finally ligated and transformed as well this week. Confirmation PCR showed bands of about 3000 bp which is consistent with the size of the operon. By Friday the successful colonies had been grown up and miniprepped, ready to send for sequencing early next week. The carAc/B0034 construct was also successfully transformed this week and verified by running a PCR using the B0034 forward primer. It was also miniprepped and is ready to send for sequencing next week.
In addition to this work constructing biobricks we also started an experiment to demonstrate the function of the pCAR plasmid in LD2. Strains of LD2 were grown up in LB and then washed and placed in B-N media + glucose and various experimental additions. These additions included carbazole, 4-PBAH (a nitrogen containing naphthenic acid), ammonia chloride, and a negative control with no nitrogen source. Strains of E. coli were also grown under the same conditions to compare growth and ammonia production of LD2 to a strain that does not contain the pCAR plasmid. These results should act as a preview of what we expect our final biobrick containing E. coli species to produce as well as allowing us to establish LD2 as a positive control for this experiment.
Week 16 (August 13 - August 17)
The sequencing for the carAc/B0034 construct came back with only carAc and no evidence of a B0034 site preceding it. The confirmation PCR that was run last week using the B0034 forward primer seemed to indicate that it was inserted, however since B0034 is only 12 base pairs the primer may not have been specific enough. The construction on this part was restarted this week. carAa colony PCR on last week's transformation showed two positive colonies, however upon culturing, miniprepping, and digesting these colonies the plasmid appeared to be too large (3000 base pairs as opposed to the expected 2000). This may be due to a problem of a mislabeled plasmid tube that has been experienced by other team members, causing the gene to be inserted into pSB1AC3 rather than pSB1C3. To test this, a streak plate was made from these colonies and grown on an ampicillin resistant plate. Mutagenized carAd was successfully transformed into Top 10 E. coli cells, cultured, and miniprepped. The miniprep product was digested with NotI to test whether the NotI site was successfully removed. These results were good, as the plasmid ran at 2000 base pairs and the insert at just under 1000 base pairs in the mutagenized carAd whereas the non-mutagenized carAd had its insert cut into 2 bands. This will be sent for sequencing next week to confirm the mutation. Unfortunately the sequencing results came back negative for the ant operon meaning we will need to restart genomic PCR on it next week. Multiple transformations and confirmation PCRs were run on AmdA this week, however none displayed a band size of 1700-1800 base pairs as expected. We have continued to try different digested plasmids as other team members also had difficulties this week with ligation into pSB1C3, possibly indicating a problem with our Antarctic Phosphatase stock.
The other news from this week was the final results from our carbazole degradation experiment attempting to demonstrate LD2's ability to degrade carbazole and hopefully other nitrogen containing compounds. Unfortunately, these results were very difficult to make sense of as the LD2 that were cultured with carbazole grew, but did not show elevated ammonia levels which would be indicative of carbazole degradation. One of our controls was LD2 cultured with ammonium chloride to test whether LD2 was capable of using ammonia as a nitrogen source. This would pose a problem for us as we were planning to use an increase in ammonia levels as a proxy for carbazole degradation rather than measuring carbazole degradation directly using GC-MS. However, the LD2 showed significantly less ammonia after 44 hours when compared to 26 hours indicating that it may be degraded, either by the LD2 or by an unaccounted for abiotic process. This has lead us to decide to use GC-MS and directly measure carbazole levels in our sample the next time we do this experiment.
Week 17 (August 20 - August 24)
The carAa was re-streaked onto ampicillin plates and grew, probably showing that it was ligated into pSB1AC3, not pSB1C3, which explained the irregularity of our confirmation digest last week. It was sent for sequencing, but in case it was incorrect we also started a new carAa pipeline by PCRing it from the Pseudomonas genome again and redigesting and ligating. It was transformed into pSB1C3 over the weekend. carAc did not see a lot of progress this week as we spent much of the time attempting to construct a RBS (B0034) in front of it, but were unsuccessful. carAd was also subjected to this construction with B0034 and we were able to send it for sequencing by the end of the week. Several new constructs were also started for carAd in case the sequencing is negative. The ant operon was successfully transformed and confirmed by colony PCR this week, after which it was grown up and sent for sequencing. We also began to attempt mutagenesis on one of the EcoRI sites in the operon, but had no amplification of the plasmid. We also re-digested some PCR purified product to start another ligation in case sequencing failed. After many attempts we were also able to finally successfully transform an AmdA/PSB1C3 plasmid into Top 10 cells this week. After a miniprep they showed bands of about 1800 base pairs on the confirmation digest gel, so they were also sent for sequencing this week and had mutagenesis started on the illegal SpeI site. Unfortunately, the sequencing sent early in the week came back with ambiguous results. The forward reaction for antABC was a match, but its reverse reaction matched to carAa. Both carAa reactions matched nothing. Since these two genes are over 20 000 base pairs apart on the template plasmid it is virtually impossible that they were actually combined into one plasmid (PCR extension time would not have been long enough and even if it was the product would have run much above the 3000 base pair mark). This is possibly a mistake on Eurofins end, so we are not 100% sure if we have 1 good gene, both of them, or neither. Also the sequence for mutagenized carAd showed only about the first 100 base pairs, which were correct however not long enough meaning it was a poor reaction and inconclusive. Finally, we also re-started our carbazole degradation assay using the GC-MS to quantitatively measure the loss of carbazole at biological sciences. This process will take 2 weeks to demonstrate LD2's ability to degrade carbazole. We are forcing it to use carbazole as a carbon and a nitrogen source in one sample and as only a nitrogen source in another sample (this sample has had glucose added to it).
Week 18 (August 27 - August 31)
The new stock of carAa that we had PCRed from LD2 was sent for sequencing this week and came back as a match, so we have begun constructing a RBS site and a TetR promoter (J13002) in front of it. At this point all the car genes have been successfully amplified, inserted into a biobrick plasmid, and sequence verified. We also continued to attempt to construct carAc with an RBS site (B0034) in front of it, but had no success. On a better note, we confirmed this week that carAd was mutated properly and we were also able to insert an RBS site in front of it. The next step for this gene will be to attempt a plasmid switch as it is currently in pSB1A3 and needs to be in pSB1C3 for submission to the parts registry. After sending a new batch of the ant operon for sequencing we got the same confusing results as we did last week. We therefore hypothesized that our primers were annealing to a homologous region that is similar to the ant genes and very close to carAa. This means that we need to restart PCR on this gene as all of our current product is incorrect. The sequencing results for AmdA that came back this week were also somewhat confusing as they did not quite match the sequence we expected. However, they were a 100% match for the AmdA gene in a different strain of Rhodococcus erythropolis. This means that DSMZ probably sent us the wrong strain of bacteria and that we were very lucky that the primers we designed based off of the expected template actually worked to amplify a slightly different gene in another strain. The good news is that this new sequence contains no illegal cut sites, which explains why mutagenesis was not working (there was nothing to mutate) and allows us to immediately begin construction of J13002 in front of AmdA this week.
Week 19 (September 3- September 7)
carAd/B0034 was successfully plasmid switched into pSB1C3, making it ready for submission. Construction of J13002 and carAa may have been successful as the digest of the miniprep looked promising. It will be sent for sequencing next week. Unfortunately there was no progress made on the constructions of AmdA with J13002 or carAc with B0034. These will be redigested and religated for next week. We could not amplify the ant operon despite trying many variations of PCR using high fidelity KAPA polymerase. This is probably due to the multiple, large regions of homology on the operon that the reverse primer binds to making amplification difficult. Because the function of this gene can theoretically be replaced by AmdA, we may scale back efforts dedicated to it, as the chances of successful amplification appear slim.
Weeks 20-22 (September 10 - September 30)
Constructs that we continued to work on this week included AmdA with J13002, carAa with J13002, and carAc with B0034. Some sequencing was sent, but no positive results were obtained. A new approach to biobrick the ant operon was implemented this week. A new forward primer was designed that would anneal about 1500 base pairs upstream of the ant operon, avoiding the region of homology that is repeated at various points on the plasmid. Once this region is successfully amplified we could use this amplified DNA as a template for another PCR reaction which would use the normal ant primers. Since the template would no longer include regions of homology these primers should be free to work properly and amplify the ant operon. The PCR using the new primers was started this week, and although amplification at 4500 base pairs was observed as expected, there was also a lot of non-specific amplification. This means that to purify the amplified DNA that we want and avoid the other products a dreaded gel extraction had to be performed which, as usual, did not work. At this point it was decided that further attempts to amplify the ant operon would be futile and our efforts would be better served working on constructing the parts we do have. Unfortunately no progress was made in these weeks on construction. We have also obtained GC-MS readings from the carbazole degradation experiment that showed that Pseudomonas LD2 can indeed degrade carbazole, however only under low glucose conditions.
Weeks 23-26 (October 1-October 24)
Most of this time was spent attempting the same construction of AmdA with a TetR promoter as described above. After this construct was finished, we designed two experiments to characterize this biobrick. One was an in vivo experiment in which E. coli cells containing the biobrick were assayed with 3 compounds containing a primary amine or amide. Benzamide, which has similar structure to the naphthenic acids being targeted by other parts of the hydrocarbon project, and butyramide were previously shown to be good substrates for the AmdA enzyme (Hirrlinger et al, 1996). Benzamidine was also tested to explore the array of substrates that could possibly be degraded by the enzyme. The other approach we took was to create a cell lysate from E. coli that was grown up and resuspended it in a lysis buffer. This was assayed with the same 3 compounds to guard against the possibility that the in vivo system lacked the appropriate transfer proteins. Hopefully this stuff works.The data below from the GC shows that one compound gets completely degraded by the cells containing the AmdA gene as well as a new product being formed in this group. The MS data shows that the compound being obliterated by amidase is in fact benzamide and that the new product being formed is benzoic acid. Benzoic acid results from replacing the NH2 with an OH group, which is the expected function of the AmdA enzyme. The protocol for this experiment can be found here
 "
"