Team:Calgary/Project/OSCAR/Denitrogenation
From 2012.igem.org
Emily Hicks (Talk | contribs) |
Emily Hicks (Talk | contribs) |
||
| Line 16: | Line 16: | ||
<h2>How Are We Going To Accomplish This?</h2> | <h2>How Are We Going To Accomplish This?</h2> | ||
| - | <p>We identified a <i>Pseudomonas</i> genus that is capable of converting carbazole into cathecol with the hope that it would also be able to act on similar nitrogen heterocycles. This is accomplished thorugh an enzymatic pathway containing carbazole-1,9-dioxygenase (<i>carA</i>) and an enzyme called anthranilate 1,2-dioxygenase. <i>carA</i> is a 4 part enzyme encoded by three genes: carA, carB and carC. It selectively cleaves the first C-N bond in a nitrogen containing ring structure, converting carbazole into 2'-aminobiphenyl-2,3-diol | + | <p>We identified a <i>Pseudomonas</i> genus that is capable of converting carbazole into cathecol with the hope that it would also be able to act on similar nitrogen heterocycles. This is accomplished thorugh an enzymatic pathway containing carbazole-1,9-dioxygenase (<i>carA</i>) and an enzyme called anthranilate 1,2-dioxygenase. <i>carA</i> is a 4 part enzyme encoded by three genes: carA, carB and carC. It selectively cleaves the first C-N bond in a nitrogen containing ring structure, converting carbazole into 2'-aminobiphenyl-2,3-diol (Xu et al, 2006,Morales, 2010). After this step, anthranilate 1,2-dioxygenase enzyme, encoded by the <i>ant</i> operon removes nitrogen from the structure (Diaz, 2010). </p>. |
</html>[[File:CarA pathway Calgary12.jpg|thumb|420px|center|Action of the <i>carA</i> enzyme complex.]]<html> | </html>[[File:CarA pathway Calgary12.jpg|thumb|420px|center|Action of the <i>carA</i> enzyme complex.]]<html> | ||
| - | <p> | + | <h2> Does it work? </h2> |
| + | |||
| + | <p>Upon obtaining the carbazole-degrading pseudomonas strain, we wanted to validate that this approach was valid. We performed an initial carbazole degradation assay where we incubated a <i>pseudomonas</i> culture with carbazole and monitored degradation over time. Our detailed protocol can be found <p><a href="https://2012.igem.org/Team:Calgary/Notebook/Protocols/carbazole">here.</a></p> | ||
| + | |||
| + | and perfomred an initial assy to show that it worked. | ||
| Line 40: | Line 44: | ||
<h2>What Have We Shown So Far?</h2> | <h2>What Have We Shown So Far?</h2> | ||
| - | |||
</html> | </html> | ||
}} | }} | ||
Revision as of 06:58, 3 October 2012


Hello! iGEM Calgary's wiki functions best with Javascript enabled, especially for mobile devices. We recommend that you enable Javascript on your device for the best wiki-viewing experience. Thanks!
Denitrogenation

Why Focus On Nitrogen Bioremediation?
The removal of nitrogen heterocycles from the petroleum tailings ponds is important for both of the main goals of the OSCAR project. Since nitrogen containing compounds lower the quality and stability of fuel (Katzer & Sivasubramanian, 1979) , their presence would be detrimental to the usefulness of the hydrocarbons OSCAR could create. The presence of nitrogen compounds in our products also hinders our environmental goals as they have been shown to undergo radical changes, yielding highly genotoxic byproducts (Xu et al., 2006). The combination of these two factors makes the removal of nitrogen very important to the success of OSCAR.
Our Model Compounds
Carbazole was chosen as the model compound to study nitrogen containing heterocycles as it accounts for about 70% of nitrogen by mass in petroleum tailings ponds, as well as the fact that it is considered one of the more difficult to degrade (Morales, 2010). A pathway that is capable of degrading carbazole could also have the ability to degrade many other types of nitrogen containing compounds found in the tailings ponds.
How Are We Going To Accomplish This?
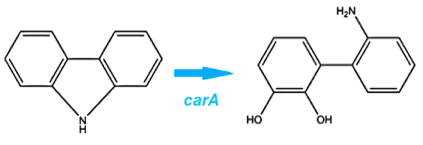
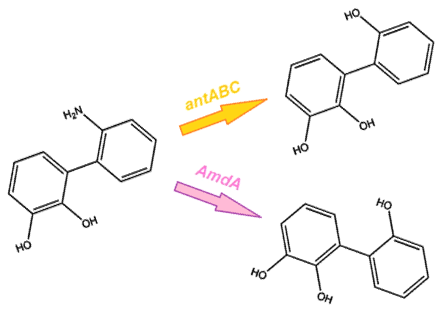
We identified a Pseudomonas genus that is capable of converting carbazole into cathecol with the hope that it would also be able to act on similar nitrogen heterocycles. This is accomplished thorugh an enzymatic pathway containing carbazole-1,9-dioxygenase (carA) and an enzyme called anthranilate 1,2-dioxygenase. carA is a 4 part enzyme encoded by three genes: carA, carB and carC. It selectively cleaves the first C-N bond in a nitrogen containing ring structure, converting carbazole into 2'-aminobiphenyl-2,3-diol (Xu et al, 2006,Morales, 2010). After this step, anthranilate 1,2-dioxygenase enzyme, encoded by the ant operon removes nitrogen from the structure (Diaz, 2010).
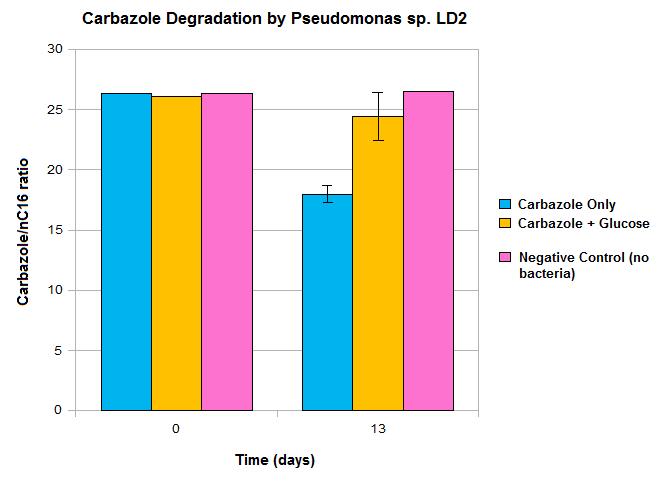
.Does it work?
Upon obtaining the carbazole-degrading pseudomonas strain, we wanted to validate that this approach was valid. We performed an initial carbazole degradation assay where we incubated a pseudomonas culture with carbazole and monitored degradation over time. Our detailed protocol can be found
and perfomred an initial assy to show that it worked.Our Constructs
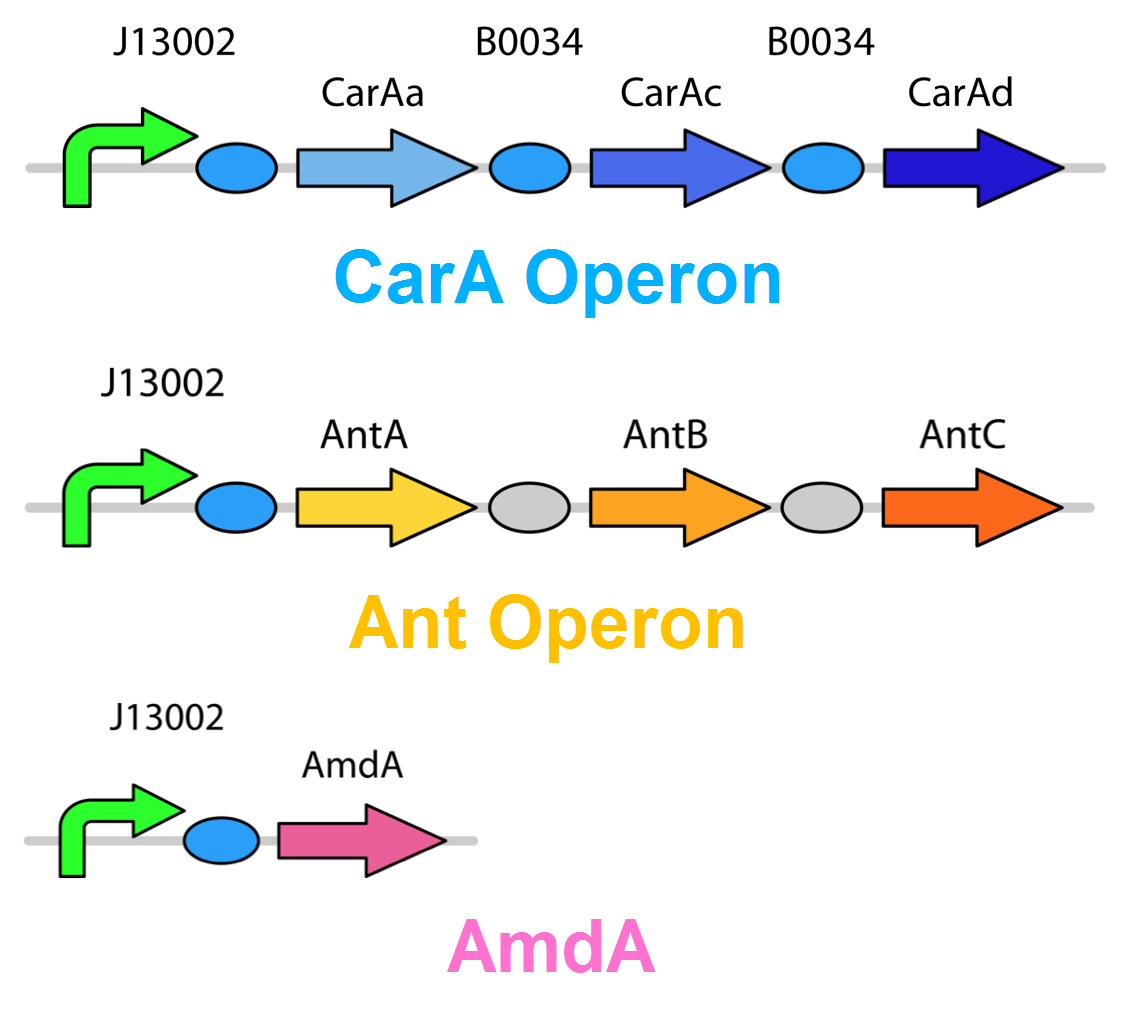
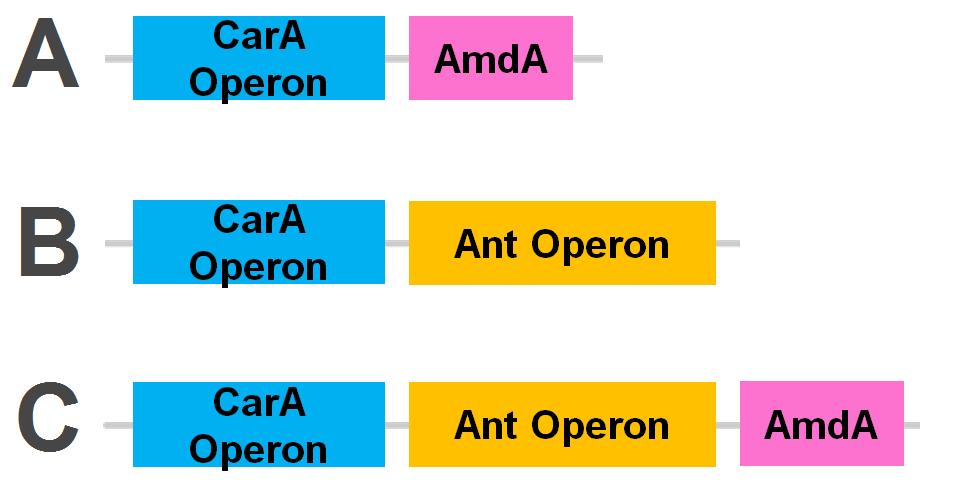
The carA genes have been biobricked individually into an operon under the control of a TetR promoter (BBa_J13002), with ribosome binding sites (BBa_B0034) inserted in front of each gene. The carAd gene was given a silent mutation to eliminate an illegal NotI site in the native sequence. The ant genes were biobricked as a whole operon with native ribosome binding sites in between each gene, also downstream of a TetR promoter. The AmdA gene was also biobricked downstream of a TetR promoter and ribosome binding site.
Using these constructs above 3 test circuits can be built. The carA operon is necessary for all three since it is the only way of accomplishing the first step in the pathway, however the AmdA and ant genes can both be used for the second step. This allows the construction of circuits which can independently test the ability of both systems to perform the final step of the pathway, as well as testing them together to see if having two pathways available makes the system significantly more efficient.
What Have We Shown So Far?
 "
"