Team:Calgary/Project/HumanPractices/Killswitch
From 2012.igem.org


Hello! iGEM Calgary's wiki functions best with Javascript enabled, especially for mobile devices. We recommend that you enable Javascript on your device for the best wiki-viewing experience. Thanks!
A Killswitch for Increased Security
Hello! We are sorry to say that this page has yet to be created. Please check back shortly to see any new updates that we may have.
Under Construction!
Currently this wiki is under construction. We hope to bring the entire site running up to full speed as soon as we can. For the time being, there will be broken links and navigation issues until we iron out the kinks. We apologize for any inconvenience.
Contents |
Purpose:
Synthetic biology entails designing an organism to do a specific task. This involves genetic manipulation and requires scientists to provide the bacteria with a selective advantage such as an antibiotic cassette which forces the bacteria to keep the gene of interest inside the cell. With such manipulation comes a valid “risk of accidental release” (Tucker and Zilinkas, 2006). In order to prevent such a bacteria from becoming rogue a killswitch is designed such that the bacteria is only able to survive in specific environments allowing them to perform the tasks of decarboxylation, denitrification and desulfurization in our bioreactor. However, in case of these bacteria escaping, the lack of a metabolite and or the presence of a particular metabolite will activate the “kill genes” which will cause the bacteria to self destruct. The killswitch mechanism was put in our system as a safety measure in addition to the bioreactor to contain the synthetic bacteria.
History:
Scientists have been trying to develop methods to limit bacterial viability and growth outside of the lab environment. One of the most popular methods used to ensure the safety of bacteria used in the lab was the creation of lab strain bacteria such as DH5α and Top10. These bacteria are metabolically deficient and are unable to survive outside of the lab environment without very specific nutrients. Additionally, The Registry of Biological Parts also has several killswitches readily available that were submitted by previous iGEM teams.
The different types of killswitches include:
Inducible kill genes:
Inducible systems generally consist of a regulatory element such as a promoter which is activated in the presence or absence of a metabolite. GIVE EXAMPLES
Toxin-antitoxin systems:
These systems usually insert antitoxin in the plasmid and toxin in the genome. Ideally if the bacteria lose the plasmid then the bacteria dies. This does not address the problem of horizontal gene transfer between species.
Auxotrophic marker
Design considerations:
During the first phase of design we considered classic systems such as auxotrophic markers, toxin-antitoxin systems, inducible systems. However, considering the cost of the system if auxotrophic markers were used we did not pursue that route. We have decided to use the inducible systems. We explored four different inducible systems which are induced by inexpensive ligands such as magnesium, manganese, molybate salts and glucose. In order to make sure the systems are controlled well and the kill switch regulation is not leaky, we have added an additional control using the riboswitch.
A riboswitch provides post-transcriptional control of gene expression. A riboswitch is a small stretch of mRNA which binds to a ligand which increases or decreases the expression of the gene downstream.
INSERT IMAGE OF RIBOSWITCH
Our kill genes:
The kill genes used in this project are S7 endonuclease from Staphylococcus aureus and a type II restriction enzyme from chlorella virus PCBV-1 called CviAII.
S7 endonuclease
S7 endonuclease is the protein product of the gene nucA from staphylococcus aureus. This endonuclease was selected due to its ability to function in low temperatures. Due to the weather in Alberta, we chose an enzyme which is able to function at low temperatures. S7 nuclease is known to function at temperature of zero degrees. FIND CITATION. Micrococcal nuclease is cleaves A-T rich regions rather than G-C rich regions (Dingwall et al, 1981). Another advantage of using this gene is the associated kinetics. This enzyme is shown to degrade genomes in less than an hour (FIND REFERENCE).
CviAII
is a restriction enzyme which cleaves DNA at the sequence CATG. TALK ABOUT KINETICS, TALK ABOUT WHY THIS WILL PROVIDE AN ADVANTAGE
Nuclease assay to evaluate the nucleases present in the registry (BglII and BamHI):
Regulation of our kill genes:
Glucose repression:
This system entails the use of a promoter called rhamnose promoter (pRha). The rhamnose promoter is repressed in presence of glucose (≥0.2%), has leaky expression in the absence of glucose and is highly upregulated in the presence of a sugar called rhamnose. In this system, the presence of rhamnose activates expression of rhaR which increases in turn expresses rhaS and rhaR. Finally, rhaS binds to pRHA in the presence of rhamnose and causes expression of the rhaBAD operon. PUT GRAPH. This system will be suppressed in the bioreactor which will contain bacterial growth medium containing glucose. This will suppress the expression of kill genes and allow our cells to perform their respective functions such as decarboxylation, denitrification and desulfurization. Ideally, if the bacteria do escape from the bioreactor into the tailings pond the lack of glucose would activate the expression of the kill genes which would destroy the bacterial genome stopping it from not only propagating in the environment but also passing on genes to other bacteria in the environment.
Magnesium regulation
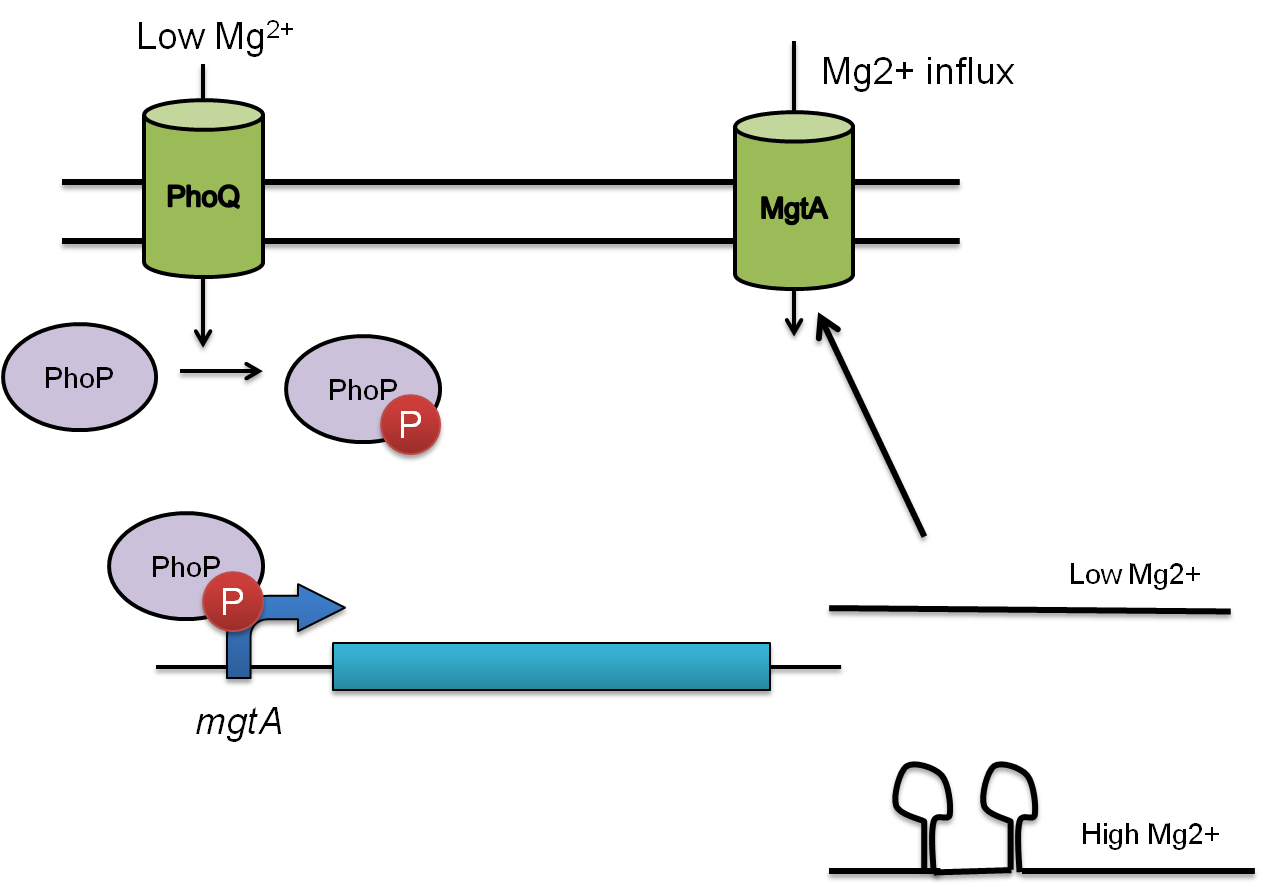
This system is repressed in the presence of magnesium. This system has two control components – a promoter and a riboswitch. Normally the magnesium promoter (mgtA promoter) and the magnesium riboswitch (mgtArb) are activated if there is a deficiency of magnesium in the cell. The lack of magnesium normally activates other genes in E. coli to bring in more magnesium into the cell. There are two proteins in the cascade that activate the system namely PhoP and PhoQ. PhoQ is the trans-membrane protein which gets activated in the absence of magnesium and phosphorylates PhoP. PhoP in turn binds to the mgtA promoter and transcribes genes downstream.
</html>
Figure 2: MgtA pathway in E. coli. The phoQ protein is the transmembrane receptor which detects low magnesium concentration. PhoQ then phosphorylates PhoP which acts as a transcription factor on mgtA promoter and transcribes genes downstream necessary for bringing magnesium into the cell. There is a second level of control with the magnesium riboswitch. In the presence of high magnesium the riboswitch forms a secondary structure which does not allow the ribosome to bind to the transcript inhibiting translation. In the case of low magnesium however, the transcript is expressed and this allows influx of magnesium.
Test circuits for the magnesium system
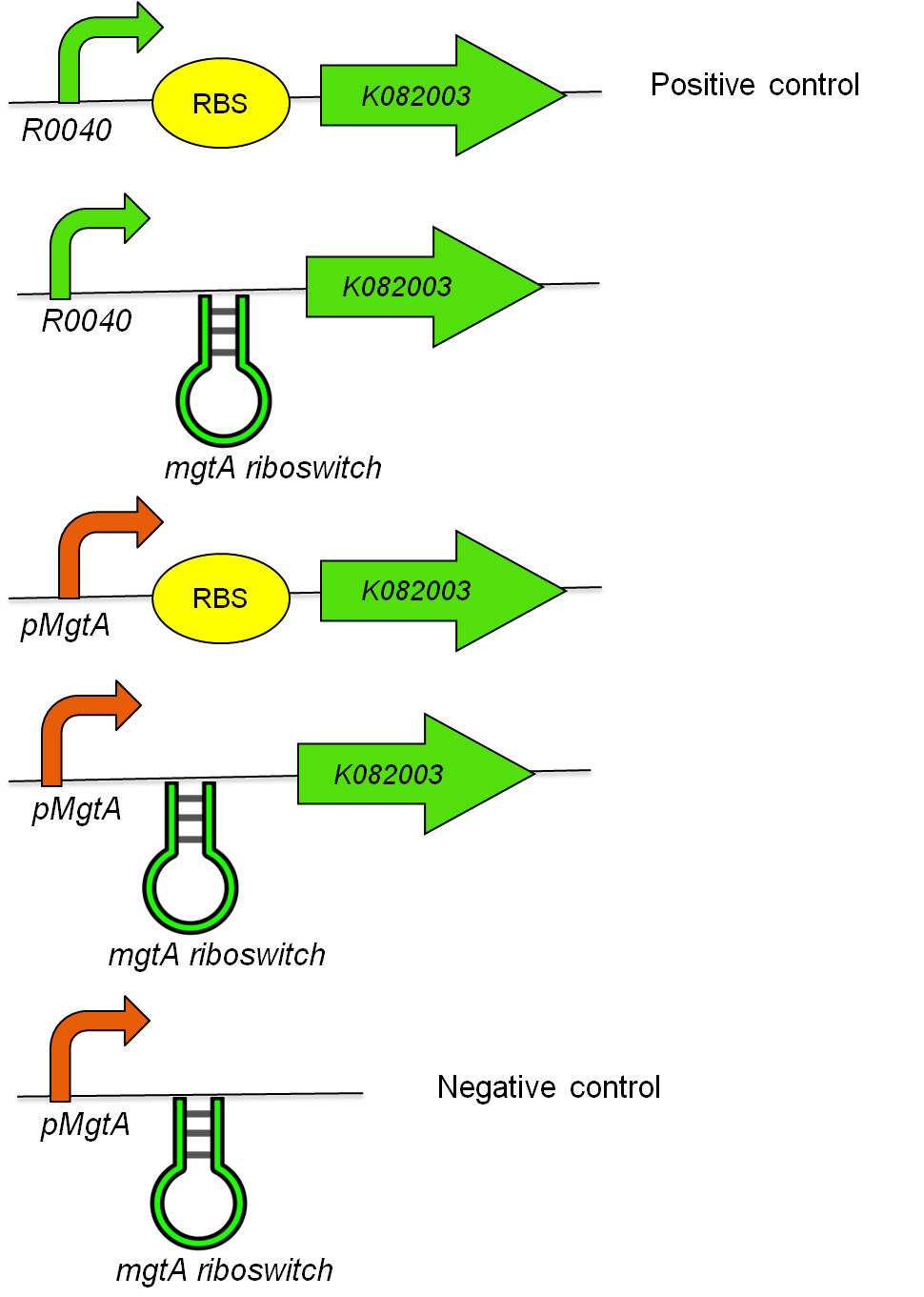
To test the magnesium regulatory elements we built each of the elements with a reporter gene. We chose Bba_K082003 which is GFP with an LVA tag as our choice of reporter. We did not choose BBa_E0040 which is the stable GFP because we wanted a real time indication of the system's control. Stable GFP has a half life of 8 hours and would still fluoresce when the system is shut off.
We build these circuits to test the control elements of the system, namely the mgtA promoter and the mgtA riboswitch.
characterization of these circuits
We tested the aforementioned circuits in different concentrations of magnesium. The values were normalized to the negative control which is the magnesium promoter and riboswitch alone.
Manganese regulation
Similar to the magnesium system, the manganese system engages both a promoter and a riboswitch. However this system operates in a reciprocal manner compared to the magnesium system. Therefore when manganese is present in the system will allow expression of the system downstream and activate our kill genes S7 and CViAII. Additionally the manganese circuit also contains a transcriptional regulator (MntR). This is regulator containing a metal-binding domain that will in the presence of manganese repress the manganese ion transporter MntH preventing the bacteria from gaining the needed metals for survival.
Both the magnesium and manganese systems are both workable killswitch constructs however after analyzing the composition of the tailing ponds the systems will not be viable without a way to regulate these two metals in the contaminated waters.
 "
"