Team:Calgary/Project/FRED/Reporting
From 2012.igem.org


Hello! iGEM Calgary's wiki functions best with Javascript enabled, especially for mobile devices. We recommend that you enable Javascript on your device for the best wiki-viewing experience. Thanks!
A Novel Electrochemical Reporting System
For FRED to be able to tell us about the toxins he's sensing we needed a good reporter system that could function in a wide array of environments. Unfortunately the traditional fluorescent or luminescent reporters have significant drawbacks that prevent them from being useful in a tailings environment that is murky and potentially anaerobic. Due to these limitations we decided to improve upon last year's electrochemical sensor using the lacZ gene to cleave a substrate into an easily detectable analyte. Our team has developed a system that utilizes three separate reporter genes to provide a triple-output biosensor. This system overcomes traditional reporters in that it is fast, accurate, and can function in turbid environments and even in the absence of oxygen!
How does it work?
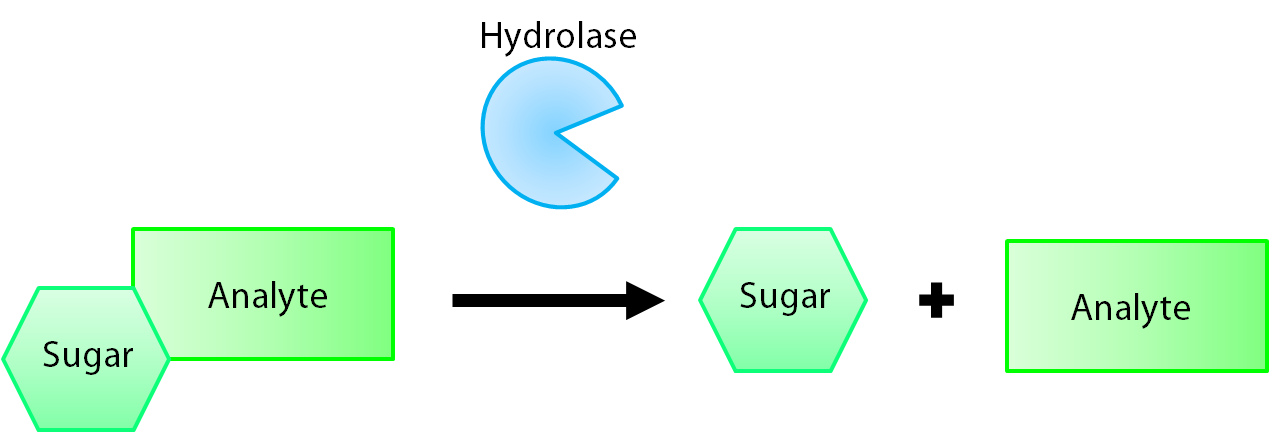
The enzymes encoded by our reporter genes are specific sugar hydrolases. This means that they target one kind of sugar and remove it from whatever compound they are attached to. We have chosen to use the sugars glucose, glucuronide, and galactose for our system. The genes reposnsible for their respective hydrolases are bglX, uidA, and lacZ. By having our electrochemical analyte conjugated to this sugar, when the hydrolase is expressed the sugar is cleaved from the analyte, allowing for it's electrochemical detection. A diagrammatic representation of this system is shown below in figure 1.
After the analyte is released we need to detect it. Electrochemistry is an excellent approach for this because of it's fast and quantitative nature. A voltage is applied between two electrodes compared to a reference electrode and the resulting current is measured. By changing the applied voltage to that of the oxidation voltage of one of our analytes, the increase in current due to it's oxidation when compared to an analyte free baseline is proportional to the amount of analyte present in the solution. This process happens so quickly that you can have an output value in a matter of seconds.
Genes, Chemicals, and Circuits
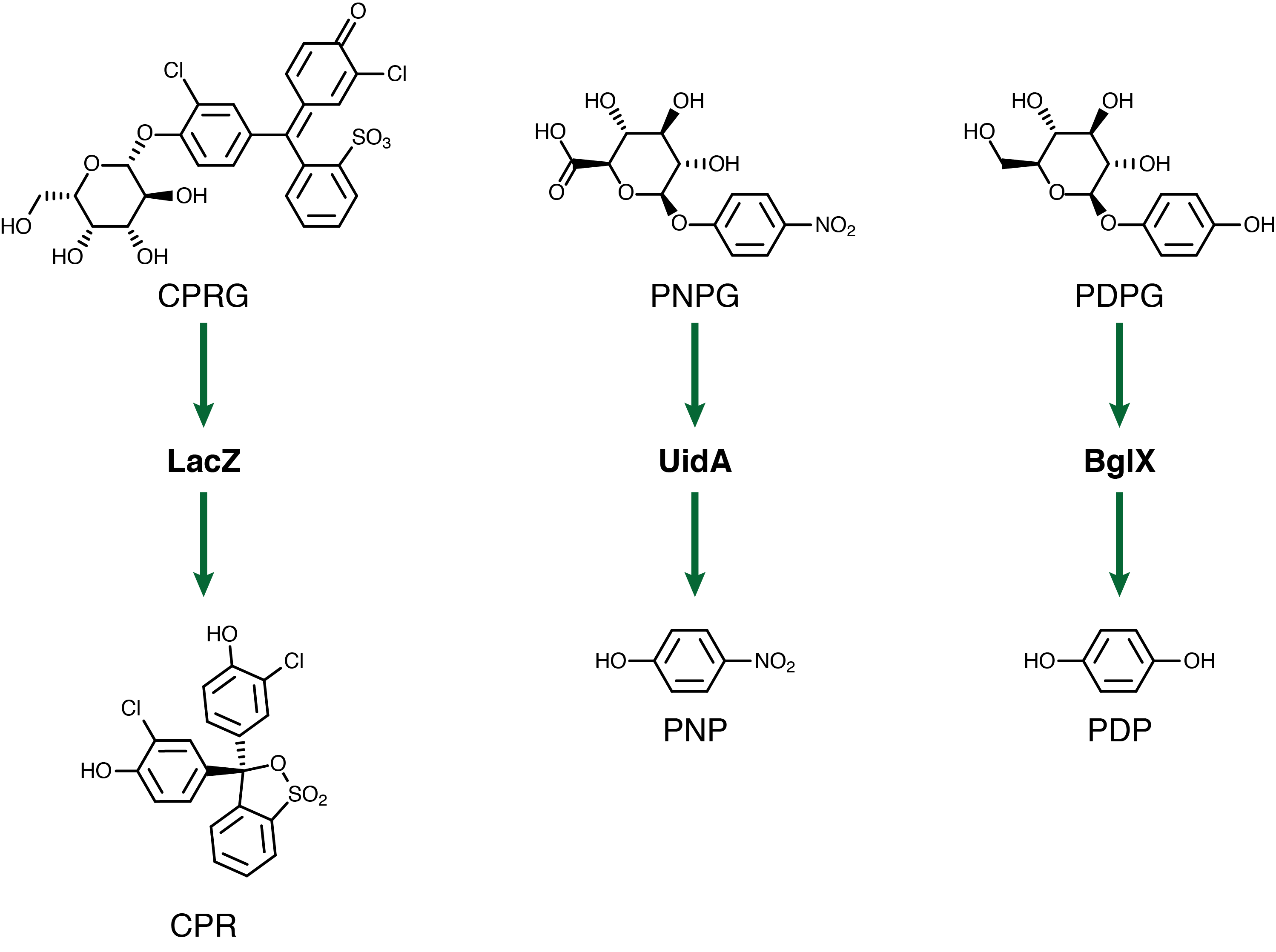
For our system to have a triplex outplut we need three seperate genetic circuits with three analytes possessing unique oxidation potentials. If one chemical overlaps with another we could get false-positives of one chemical due to oxidation of another. To this end we have chosen to use chlorophenol red (CPR), para-diphenol (PDP), and para-nitrophenol (PNP). These compounds are conjugated with their sugars to form CPR-β-D-galactopyranoside (CPRG), PDP-β-D-glucopyranoside (PDPG), and PNP-β-D-glucuronide (PNPG). An easy way to tell the analytes from their sugar conjugates is the addition of the letter G to the acronym. These chemicals are summarized below in figure 2 along with the reporter genes used with each one.
Out of the three sugar conjugates the only one that exhibits any electrochemical activity is PDPG, with it's oxidation potential at 0.6V vs the reduction of hydrogen reference electrode (RHE). The three analytes have potentials at 0.825V for PDP, 1.4V for CPR, and 1.6V for PNP vs RHE. As none of these peaks overlap and no sugar conjugates interfere with their signals the three chemicals can be detected in the same solution. This is shown below in figure 3.
CV OF ALL 3 ANALYTES AT ONCE
With the chemicals finalized we now needed to construct our circuits. The lacZ gene under the control of the lacI promoter in the registry has a frameshift mutation, rendering the enzyme nonfunctional. As such we have reconstructed this circuit and submitted it to the registry (BBa_K902008???). The bglX and uidA genes were amplified from the E. coli genome using PCR and biobricked as BBa_K902004 and BBa_K902000 respectively. These genes were then also constructed under the lacI promoter to allow for comparison testing.
 "
"