Team:OUC-China/Project/DesignMaking/GoalandDesign
From 2012.igem.org
Zhangpeiran (Talk | contribs) |
|||
| (17 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
<script type="text/javascript" src="http://1.oucigem.sinaapp.com/OUC-nav.js"></script><!--nav js--> | <script type="text/javascript" src="http://1.oucigem.sinaapp.com/OUC-nav.js"></script><!--nav js--> | ||
<link rel="stylesheet" href="http://1.oucigem.sinaapp.com/style.css"><!--slidenav css--> | <link rel="stylesheet" href="http://1.oucigem.sinaapp.com/style.css"><!--slidenav css--> | ||
| - | + | <script type="text/javascript" src="http://1.oucigem.sinaapp.com/zhangpeiran-jquery.easing.1.3.js"></script> | |
| - | <style> | + | <style type="text/css"> |
| + | <!-- | ||
| + | body{margin:0px; padding:0px; background:#fff;} | ||
| + | #fl_menu{position:absolute; top:50px; left:1127px; z-index:9999; width:150px; height:50px;} | ||
| + | #fl_menu .label{padding-left:20px; line-height:50px; font-family:"Arial Black", Arial, Helvetica, sans-serif; font-size:14px; font-weight:bold; background:#000; color:#fff; letter-spacing:7px;} | ||
| + | #fl_menu .m2enu{display:none;} | ||
| + | #fl_menu .m2enu .menu_item{display:block; background:#000; color:#bbb; border-top:1px solid #333; padding:10px 20px; font-family:Arial, Helvetica, sans-serif; font-size:12px; text-decoration:none;} | ||
| + | #fl_menu .m2enu a.menu_item:hover{background:#333; color:#fff;} | ||
| + | .content{width:520px; margin:50px auto;} | ||
| + | --> | ||
| + | #back-top { | ||
| + | position: fixed; | ||
| + | bottom: 60px; | ||
| + | margin-left: 1160px; | ||
| + | } | ||
| + | #back-top a { | ||
| + | width:85px; | ||
| + | display: block; | ||
| + | text-align: center; | ||
| + | font: 11px/100% Arial, Helvetica, sans-serif; | ||
| + | text-transform: uppercase; | ||
| + | text-decoration: none; | ||
| + | color: #bbb; | ||
| + | -webkit-transition: 1s; | ||
| + | -moz-transition: 1s; | ||
| + | transition: 1s; | ||
| + | } | ||
| + | #back-top a:hover { | ||
| + | color: #000; | ||
| + | } | ||
| + | #back-top span { | ||
| + | width: 85px; | ||
| + | height: 85px; | ||
| + | display: block; | ||
| + | margin-bottom: 7px; | ||
| + | background: #B9B7B7 url(https://static.igem.org/mediawiki/2012/5/5e/Ouc-2012-up-arrow.png) no-repeat center center; | ||
| + | -webkit-border-radius: 15px; | ||
| + | -moz-border-radius: 15px; | ||
| + | border-radius: 15px; | ||
| + | -webkit-transition: 1s; | ||
| + | -moz-transition: 1s; | ||
| + | transition: 1s; | ||
| + | } | ||
| + | #back-top a:hover span { | ||
| + | background-color: #777; | ||
| + | } | ||
| + | .mini-riboon{ | ||
| + | position: relative; | ||
| + | z-index:0; | ||
| + | left: 1001px; | ||
| + | top: 90px; | ||
| + | width: 106px; | ||
| + | height: 45px; | ||
| + | } | ||
*{ | *{ | ||
margin:0; | margin:0; | ||
| Line 19: | Line 72: | ||
padding:0; | padding:0; | ||
margin:0; | margin:0; | ||
| - | overflow-x: | + | overflow-x:auto; |
font: 500 12px/1.625 "Helvetica Neue", Helvetica, Arial, sans-serif; | font: 500 12px/1.625 "Helvetica Neue", Helvetica, Arial, sans-serif; | ||
| + | } | ||
| + | #top-section{ | ||
| + | width:99%; | ||
} | } | ||
#page{ | #page{ | ||
| Line 139: | Line 195: | ||
text-align:center; | text-align:center; | ||
margin-top:25px; | margin-top:25px; | ||
| - | margin-left: | + | margin-left:10px; |
| + | width:800px; | ||
} | } | ||
| Line 145: | Line 202: | ||
height: 50px; | height: 50px; | ||
| - | width: | + | width:1349px; |
margin-top:0; | margin-top:0; | ||
margin-bottom:-10px; | margin-bottom:-10px; | ||
background-color:#1487DF; | background-color:#1487DF; | ||
| - | + | padding-left:0; | |
| - | + | padding-right:0; | |
| - | border | + | border:none; |
} | } | ||
#footer a | #footer a | ||
| Line 186: | Line 243: | ||
</div> | </div> | ||
<ul class="menu"> | <ul class="menu"> | ||
| - | <li> <a href="https://2012.igem.org/Team:OUC-China"> | + | <li> <a href="https://2012.igem.org/Team:OUC-China">Index</a> </li> |
<li> <a href="https://2012.igem.org/Team:OUC-China/Project/Overview">Project</a> </li> | <li> <a href="https://2012.igem.org/Team:OUC-China/Project/Overview">Project</a> </li> | ||
<li> <a href="https://2012.igem.org/Team:OUC-China/Modeling/Overview">Modeling</a> </li> | <li> <a href="https://2012.igem.org/Team:OUC-China/Modeling/Overview">Modeling</a> </li> | ||
| Line 199: | Line 256: | ||
<ul class="submenu"> | <ul class="submenu"> | ||
<li> | <li> | ||
| - | <a href="https://2012.igem.org/Team:OUC-China | + | <a href="https://2012.igem.org/Team:OUC-China"> |
<img src="https://static.igem.org/mediawiki/2012/a/af/Projectoverview.jpg" /> | <img src="https://static.igem.org/mediawiki/2012/a/af/Projectoverview.jpg" /> | ||
| - | + | Home | |
</a> | </a> | ||
</li> | </li> | ||
<li> | <li> | ||
| - | <a href="https://2012.igem.org/Team:OUC-China | + | <a href="https://2012.igem.org/Team:OUC-China#hmo"> |
<img src="https://static.igem.org/mediawiki/2012/4/49/Humanpracticeoverview.jpg" /> | <img src="https://static.igem.org/mediawiki/2012/4/49/Humanpracticeoverview.jpg" /> | ||
| - | HumanPractice | + | HumanPractice index |
</a> | </a> | ||
</li> | </li> | ||
| Line 219: | Line 276: | ||
<a href="https://2012.igem.org/Team:OUC-China/contactus"> | <a href="https://2012.igem.org/Team:OUC-China/contactus"> | ||
<img src="https://static.igem.org/mediawiki/2012/1/1e/Contact-us.jpg" /> | <img src="https://static.igem.org/mediawiki/2012/1/1e/Contact-us.jpg" /> | ||
| + | Contact Us | ||
</a> | </a> | ||
| - | + | ||
</li> | </li> | ||
</ul> | </ul> | ||
| Line 317: | Line 375: | ||
<a href="https://2012.igem.org/Team:OUC-China/Team/AcknowledgementandCooperation"> | <a href="https://2012.igem.org/Team:OUC-China/Team/AcknowledgementandCooperation"> | ||
<img src="https://static.igem.org/mediawiki/2012/a/a6/Ouc-nav-co.jpg" /> | <img src="https://static.igem.org/mediawiki/2012/a/a6/Ouc-nav-co.jpg" /> | ||
| - | + | Acknowledgement&Cooperation | |
</a> | </a> | ||
</li> | </li> | ||
| Line 359: | Line 417: | ||
<a href="https://2012.igem.org/Team:OUC-China/protocol"> | <a href="https://2012.igem.org/Team:OUC-China/protocol"> | ||
<img src="https://static.igem.org/mediawiki/2012/7/7d/Ouc-pro.png" /> | <img src="https://static.igem.org/mediawiki/2012/7/7d/Ouc-pro.png" /> | ||
| - | + | Protocols | |
</a> | </a> | ||
</li> | </li> | ||
| Line 366: | Line 424: | ||
<!-- content --> | <!-- content --> | ||
<div id="page"> | <div id="page"> | ||
| + | <div id="fl_menu"> | ||
| + | <div class="label">MENU</div> | ||
| + | <div class="m2enu"> | ||
| + | <a href="https://2012.igem.org/Team:OUC-China/Project/DesignMaking/GoalandDesign#goal" class="menu_item">Goal</a> | ||
| + | <a href="https://2012.igem.org/Team:OUC-China/Project/DesignMaking/GoalandDesign#design" class="menu_item">Design</a> | ||
| + | <a href="https://2012.igem.org/Team:OUC-China/Project/DesignMaking/GoalandDesign#platform" class="menu_item">Platform Construction</a> | ||
| + | <a href="https://2012.igem.org/Team:OUC-China/Project/DesignMaking/GoalandDesign#theoretical" class="menu_item">Theoretical analysis through modeling</a> | ||
| + | <a href="https://2012.igem.org/Team:OUC-China/Project/DesignMaking/GoalandDesign#rna" class="menu_item">RNA Scaffold and Platform assembly and quantitative tests</a> | ||
| + | <a href="https://2012.igem.org/Team:OUC-China/Project/DesignMaking/GoalandDesign#comparator" class="menu_item">Artificial Region Design: Comparator</a> | ||
| + | <a href="https://2012.igem.org/Team:OUC-China/Project/DesignMaking/GoalandDesign#sensor" class="menu_item">Artificial Region Design: Ratio sensor</a> | ||
| + | <a href="https://2012.igem.org/Team:OUC-China/Project/DesignMaking/GoalandDesign#region" class="menu_item">Artificial region assembly & Testing quantitatively</a> | ||
| + | </div> | ||
| + | </div> | ||
<div id="me2nu"> | <div id="me2nu"> | ||
<div id="menu_bar"> | <div id="menu_bar"> | ||
| Line 378: | Line 449: | ||
</div> | </div> | ||
</div> | </div> | ||
| + | <div class="mini-riboon"><img src="https://static.igem.org/mediawiki/2012/0/03/Ouc-mini-ribbon-decision-making.png"></div> | ||
<div class="article-list"> | <div class="article-list"> | ||
<div class="start"></div> | <div class="start"></div> | ||
<div class="list-box"> | <div class="list-box"> | ||
<div class="list-text"> | <div class="list-text"> | ||
| - | <h1>Goal</h1> | + | <h1 style="border:none;" id="goal">Goal</h1> |
<p> | <p> | ||
<a><img style="margin-top:5px;" src="https://static.igem.org/mediawiki/2012/c/c2/Ouc-project-designmaking-goal0.jpg" /></a> | <a><img style="margin-top:5px;" src="https://static.igem.org/mediawiki/2012/c/c2/Ouc-project-designmaking-goal0.jpg" /></a> | ||
| Line 395: | Line 467: | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| - | <h1>Design</h1> | + | <h1 style="border:none;" id="design">Design</h1> |
<p><strong>Insights into the “black box”</strong> | <p><strong>Insights into the “black box”</strong> | ||
<span></span>Considering the RNA pairing reaction as chemical reaction,we will see the probability of collision is determined by their concentration, and the probability of pairing after collision is determined by their affinity(that is reaction constant k in mass-action law). | <span></span>Considering the RNA pairing reaction as chemical reaction,we will see the probability of collision is determined by their concentration, and the probability of pairing after collision is determined by their affinity(that is reaction constant k in mass-action law). | ||
| Line 410: | Line 482: | ||
For Ratio Sensor,we selected a small RNA scaffold which would fuse artificial region to 5’ end of small RNA to construct a strong sRNA::sRNA interaction,the sRNA buffer with each other in cytosol in this case; | For Ratio Sensor,we selected a small RNA scaffold which would fuse artificial region to 5’ end of small RNA to construct a strong sRNA::sRNA interaction,the sRNA buffer with each other in cytosol in this case; | ||
For comparator,km2=0,which means sRNA2 is a pool of buffer for sRNA1,at the same time establish a tunable threshold for comparing quantitatively. | For comparator,km2=0,which means sRNA2 is a pool of buffer for sRNA1,at the same time establish a tunable threshold for comparing quantitatively. | ||
| - | < | + | |
| + | <a><img style="margin-left:170px;" src="https://static.igem.org/mediawiki/2012/7/7c/Ouc-project-designmaking-design1.JPG" /></a> | ||
| + | <p style="text-align:center; font-size:90%;">Fig.2 Final construct of our trial decision-making device. | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
NOTE:several independent units in this figure maybe assembly on different copy number plasmids for double-plasmids transformation | NOTE:several independent units in this figure maybe assembly on different copy number plasmids for double-plasmids transformation | ||
</p> | </p> | ||
| - | |||
<br/> | <br/> | ||
<a><img src="https://static.igem.org/mediawiki/2012/6/6b/Ouc-project_overview.png" /></a> | <a><img src="https://static.igem.org/mediawiki/2012/6/6b/Ouc-project_overview.png" /></a> | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| - | <h2>Platform Construction <a href="https://2012.igem.org/Team:OUC-China/Project/Supplementary">(experimental flow chart)</a></h2> | + | <h2 id="platform">Platform Construction <a href="https://2012.igem.org/Team:OUC-China/Project/Supplementary">(experimental flow chart)</a></h2> |
<p><span></span>We construct tet/lac_GFP generator below to characterize R0011(Plac) and R0040(Ptet) promoting behaviour,so that we could alter the sRNA levels in cytosol relatively.<a href="https://2012.igem.org/Team:OUC-China/Project/DesignMaking/Result">Results</a> | <p><span></span>We construct tet/lac_GFP generator below to characterize R0011(Plac) and R0040(Ptet) promoting behaviour,so that we could alter the sRNA levels in cytosol relatively.<a href="https://2012.igem.org/Team:OUC-China/Project/DesignMaking/Result">Results</a> | ||
| - | < | + | |
| + | <a><img style="margin-left:170px;" src="https://static.igem.org/mediawiki/2012/2/26/Ouc-project-designmaking-design2.JPG" /></a> | ||
| + | <p style="text-align:center; font-size:90%;">Fig.3 sRNA assembly platform. | ||
<br/> | <br/> | ||
<br/>E0240 is a classic GFP translational unit with weak RBS to characterize the promoter behaviour | <br/>E0240 is a classic GFP translational unit with weak RBS to characterize the promoter behaviour | ||
</p> | </p> | ||
| - | |||
<br/> | <br/> | ||
<br/> | <br/> | ||
| - | <h2>Theoretical analysis through modeling</h2> | + | <h2 id="theoretical">Theoretical analysis through modeling</h2> |
<p><strong>What’s in the 3-RNA system?</strong><span></span>We formulated the mechanism above quantitatively via a simple kinetic model(ODE model) for three RNA interaction. The model is cast in terms of two mass-action equations for the cellular concentrations of the sRNA ([s1],[s2]) and its target GFP mRNA ([m]): | <p><strong>What’s in the 3-RNA system?</strong><span></span>We formulated the mechanism above quantitatively via a simple kinetic model(ODE model) for three RNA interaction. The model is cast in terms of two mass-action equations for the cellular concentrations of the sRNA ([s1],[s2]) and its target GFP mRNA ([m]): | ||
<br/> | <br/> | ||
| Line 472: | Line 546: | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| - | <h2>RNA Scaffold and Platform assembly and quantitative tests <a href="https://2012.igem.org/Team:OUC-China/Project/Supplementary">(experimental flow chart)</a></h2> | + | <h2 id="rna">RNA Scaffold and Platform assembly and quantitative tests <a href="https://2012.igem.org/Team:OUC-China/Project/Supplementary">(experimental flow chart)</a></h2> |
<p><span></span>We ligate RNA scaffold and previous platform together ,then tested them quantitatively by plate reader, characterize the relationship between inducer concentration gradient and <i>spot42</i>-galK::GFP repression efficiency. | <p><span></span>We ligate RNA scaffold and previous platform together ,then tested them quantitatively by plate reader, characterize the relationship between inducer concentration gradient and <i>spot42</i>-galK::GFP repression efficiency. | ||
<br/> | <br/> | ||
| Line 486: | Line 560: | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| - | <h2>Artificial Region Design: Comparator</h2> | + | <h2 id="comparator">Artificial Region Design: Comparator</h2> |
<br/> | <br/> | ||
<a><img style="margin-left:8px;" src="https://static.igem.org/mediawiki/2012/5/5f/Ouc-project-designmaking-design11.JPG" /></a> | <a><img style="margin-left:8px;" src="https://static.igem.org/mediawiki/2012/5/5f/Ouc-project-designmaking-design11.JPG" /></a> | ||
| Line 515: | Line 589: | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| - | <h2>Artificial Region Design: Ratio sensor</h2> | + | <h2 id="sensor">Artificial Region Design: Ratio sensor</h2> |
<br/> | <br/> | ||
<br/> | <br/> | ||
| Line 564: | Line 638: | ||
<br/> | <br/> | ||
<br/> | <br/> | ||
| - | <h2>Artificial region assembly & Testing quantitatively</h2> | + | <h2 id="region">Artificial region assembly & Testing quantitatively</h2> |
<p><strong>Assembly method</strong> | <p><strong>Assembly method</strong> | ||
<span></span>We amplify artificial region by PCR from DH5α,using primers with ClaI/SalI site; | <span></span>We amplify artificial region by PCR from DH5α,using primers with ClaI/SalI site; | ||
| Line 629: | Line 703: | ||
</div> | </div> | ||
</div> | </div> | ||
| + | <p id="back-top"> | ||
| + | <a href="#top"><span></span>Back to Top</a> | ||
| + | </p> | ||
</body> | </body> | ||
| + | <script type="text/javascript"> | ||
| + | function FloatMenu(){ | ||
| + | var animationSpeed=1500; | ||
| + | var animationEasing='easeOutQuint'; | ||
| + | var scrollAmount=$(document).scrollTop(); | ||
| + | var newPosition=menuPosition+scrollAmount; | ||
| + | if($(window).height()<$('#fl_menu').height()+$('#fl_menu .m2enu').height()){ | ||
| + | $('#fl_menu').css('top',menuPosition); | ||
| + | } else { | ||
| + | $('#fl_menu').stop().animate({top: newPosition}, animationSpeed, animationEasing); | ||
| + | } | ||
| + | } | ||
| + | $(window).load(function() { | ||
| + | menuPosition=$('#fl_menu').position().top; | ||
| + | FloatMenu(); | ||
| + | }); | ||
| + | $(window).scroll(function () { | ||
| + | FloatMenu(); | ||
| + | }); | ||
| + | jQuery(document).ready(function(){ | ||
| + | var fadeSpeed=500; | ||
| + | $("#fl_menu").hover( | ||
| + | function(){ //mouse over | ||
| + | $('#fl_menu .label').fadeTo(fadeSpeed, 1); | ||
| + | $("#fl_menu .m2enu").fadeIn(fadeSpeed); | ||
| + | }, | ||
| + | function(){ //mouse out | ||
| + | $('#fl_menu .label').fadeTo(fadeSpeed, 0.75); | ||
| + | $("#fl_menu .m2enu").fadeOut(fadeSpeed); | ||
| + | } | ||
| + | ); | ||
| + | }); | ||
| + | </script> | ||
<script src="http://www.google-analytics.com/urchin.js" type="text/javascript"> | <script src="http://www.google-analytics.com/urchin.js" type="text/javascript"> | ||
| + | </script> | ||
| + | <script> | ||
| + | $(document).ready(function(){ | ||
| + | // hide #back-top first | ||
| + | $("#back-top").hide(); | ||
| + | |||
| + | // fade in #back-top | ||
| + | $(function () { | ||
| + | $(window).scroll(function () { | ||
| + | if ($(this).scrollTop() > 100) { | ||
| + | $('#back-top').fadeIn(); | ||
| + | } else { | ||
| + | $('#back-top').fadeOut(); | ||
| + | } | ||
| + | }); | ||
| + | |||
| + | // scroll body to 0px on click | ||
| + | $('#back-top a').click(function () { | ||
| + | $('body,html').animate({ | ||
| + | scrollTop: 0 | ||
| + | }, 800); | ||
| + | return false; | ||
| + | }); | ||
| + | }); | ||
| + | }); | ||
</script> | </script> | ||
<script> | <script> | ||
Latest revision as of 03:45, 27 October 2012

Goal
We are going to construct a trial device to process two different inputs(eg.aTc,IPTG) into detectable fluorescence change.

After constructing the trial device,we will assembly the fine-tuned Nitrate/Phosphate sensors and Gas Vesicle generator to a well characterized decision-making device,to make a full version Red Tide Alarmer.
Design
Insights into the “black box”
Considering the RNA pairing reaction as chemical reaction,we will see the probability of collision is determined by their concentration, and the probability of pairing after collision is determined by their affinity(that is reaction constant k in mass-action law).

Fig.1 Insights into the black box: “[ ]” stands for concentration; “k” stands for RNA reaction constant
Decision-making device(trial) final construct
Tunable promoter Ptet/Plac controls the transcription rate(αs,nM/min) of two small RNAs;we expect the aTc/IPTG gradient will generate different small RNA levels in cytosol,which could be monitored by our GFP generator that has been fused to sRNA target site.
For Ratio Sensor,we selected a small RNA scaffold which would fuse artificial region to 5’ end of small RNA to construct a strong sRNA::sRNA interaction,the sRNA buffer with each other in cytosol in this case;
For comparator,km2=0,which means sRNA2 is a pool of buffer for sRNA1,at the same time establish a tunable threshold for comparing quantitatively.

Fig.2 Final construct of our trial decision-making device.
NOTE:several independent units in this figure maybe assembly on different copy number plasmids for double-plasmids transformation

Platform Construction (experimental flow chart)
We construct tet/lac_GFP generator below to characterize R0011(Plac) and R0040(Ptet) promoting behaviour,so that we could alter the sRNA levels in cytosol relatively.Results

Fig.3 sRNA assembly platform.
E0240 is a classic GFP translational unit with weak RBS to characterize the promoter behaviour
Theoretical analysis through modeling
What’s in the 3-RNA system?We formulated the mechanism above quantitatively via a simple kinetic model(ODE model) for three RNA interaction. The model is cast in terms of two mass-action equations for the cellular concentrations of the sRNA ([s1],[s2]) and its target GFP mRNA ([m]):



Is it feasible?
In this three-ternary system,there are at least 6 parameters uncertain,we verified the feasibility of constructing such a device through ODE simulation using parameter set from previous researches, fortunately,it is viable theoretically within the affordable parameter range in cell,and it’s apparent enough considering stochasticity.
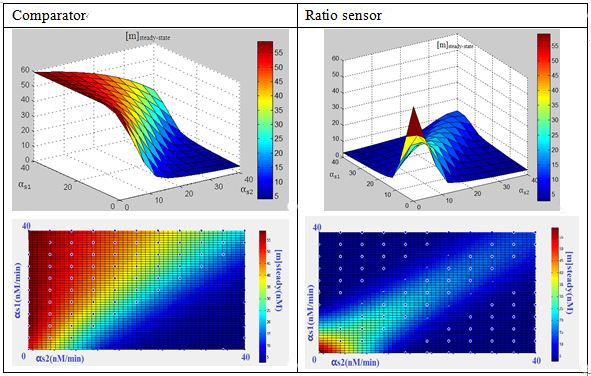
Subsquent analysisWe conducted a theoretical analysis consists of four steps(Modeling Section)

The modeling analysis shed some light on small RNA scaffold selection:in this three-ternary system,one thing is clear, km is the most important parameter in this system in given parameter set,if km is not low enough(>1),there won’t be any chance to get the ideal behaviours by fine-tuning other accessible parameters like the range of transcription rate(α) and sRNA::sRNA binding rate(ks).
sRNA scaffold selection:

Fig.4 Fold regulation was calculated from the relative fluorescence by dividing the fluorescence obtained in the presence of the sRNA plasmid by that obtained with a control plasmid that does not express a regulatory RNA.
We investigated many srNAs,finally selected spot42 sRNA and galk::mRNA as our engineering chassis for several reasons:
-1 Theoretical analysis has shown that the mRNA repression rate must be weak enough,we screened small RNAs from characterization data,found that spot42 had the weakest repression capability;
-2 spot42 is a multitarget sRNA,which is favourable in screening comparator buffer RNA with varied affinity;
-3 spot42 has a weak RNA chaperone Hfq binding site,where as galK weaker,which is favourable in sRNA::sRNA interaction with Hfq;
-4 It’s a very strong pairing(up to 20bases),but it only causes 2.6 fold repression according to Johannes H. Urban and Jo¨ rg Vogel,for it doesn’t result in the degradation of the RNA complex by RNaseE(ssRNA degradation) and RNaseIII(dsRNA degradation),both of them are major enzymes that causes RNA degradation in vivo.spot42-galK complex degrades in a slow and presently unclear way;
-5 Well studied,data is sufficient enough.
RNA Scaffold and Platform assembly and quantitative tests (experimental flow chart)
We ligate RNA scaffold and previous platform together ,then tested them quantitatively by plate reader, characterize the relationship between inducer concentration gradient and spot42-galK::GFP repression efficiency.

The modular architecture of artificial small RNA are as follows:

Artificial Region Design: Comparator

1.Modeling analysis
As we have mentioned, modeling analysis tells us that interactions between comparator buffer RNA(targets of spot42) and spot42 should be far stronger than galK::gfp transcript and spot42. To this end, we need to make in-depth investigations in spot42-mediated gene silencing.See more.
2.spot42 targets investigation
Fortunately Chase L. Beisel has made detailed works about the network that spot42 is involved in catabolite repression. Following microarray analysis and beta-galactosidase assays for LacZ-fusion reported by Chase L. Beisel, we finally selected three spot42 targets, nanC, ytfJ and srlA as our candidates for comparator buffer RNA. Here are our candidates’ results of microarray analysis.

Fig.5 The ratio of mRNA levels for pBRplac and pspot42 samples is shown at the right end. We could see that the repression rates of all our candidates are more than 2.6-fold, higher than galK. Although nanC does not work well in microarray analysis, the author indicated that nanC::lacZ fusion gives surprisingly more than 47-fold repression rate which seems to match our requirement well.Learn more.
3.Analysis secondary structure & binding sites of target mRNA
Next we considered if there are motifs that facilitate the interaction by means of some RNA prediction software such as RNA structure, intaRNA and NUPACK.
After confirming the interactive mechanisms, we decided to construct a trial platform to verify the feasibility of our design and acquire the first-hand data for further fine-tuning. The brief introduction of our comparator trial device is below.Learn more.

Fig.6 our design for double plasmids inspection.On plasmid J61002, P0440 can produce repressor TetR to repress R0040 while P0412 can produce repressor LacI to repress R0011.The Repression caused by TetR and LacI can be offset by aTc and IPTG repectively, resulting in transcription of buffer RNA and spot42. The other plasmid on the left side contains a reporter gene GFP, fusing with galK. Provided that concentration of spot42 in cytosol surpasses that of buffer RNA, it will act as galK::GFP inhibitor to repress the expression of GFP
Artificial Region Design: Ratio sensor

Modeling analysisSimilar to our comparator design, we first adjusted our design ideas according to the model we construct. It indicates that ks should be far higher than km which enables sRNA-sRNA interactions prior to sRNA-mRNA interactions.See more.
sRNA investigation
To this end, we have compared a lot of sRNA-mRNA interactions, both trans-acting and cis-acting. Unfortunately, few literatures tried to make comparison between different RNA expression rates for artificial design, let alone the construction of ternary RNAs system. So we have worked extremely hard to shape and improve our design ideas.
In silico analysis of 2 different design guidelines
First, we searched for sRNA-mRNA interactive mechanisms that show potential for relatively strong interaction. We have brainstormed several ideas in which we picked up only one that showed great potential for success. We singled out translational activated reaction of dsrA-rpos as our ‘raw materials’ for artificial region. It works like this: dsrA activates rpos translation by base-pairing with the 5’ rpos leader, which relieves an intra-molecular stem-loop structure that sequesters the rpos ribosome binding site (RBS). The location of complementary pairing site of dsrA& rpos is shown below.

Secondary structure of dsrA&rpos as well as completed artificial sRNA(dsrA::spo42 & rpos::spot42) is shown below.

While our first design to construct a strengthened inert secondary structure of sRNA-sRNA is on progress, another design gradually matured, given that we have successfully constructed comparator which is based on multi-targets spot42 network. The concept of this design focuses more on biological reaction mechanisms that blocking of RBS to prevent ribosomes loading on galK::GFP transcripts is necessary for GFP expression silencing. In view of this, it seems not reliable to simply look for non-cognated sRNA pairs to assembly to spot42 respectively.
Designs of comparator shed light on us that targets of spot42 inserted into the AR(artificial region) might solve this problem. To this end, we have chosen nanC as an attempt for it. However, it is frustrating that when nanC with various lengths were assembled with spot42 in silico, its secondary structure never failed to changed, leading to loss of functional domains. An orthogonal design finally solved this disturbing problem. Pair of mutant variant in spot42 and complementary mutant variant in nanC have been reported previously and they worked perfectly as well. So we decided to introduce 5 bases mutant in nanC(nanC*) as well as complementary mutant in spot42(*spot42). Obviously nanC* is deprived of pairing with spot42, but given ability to pair with *spot42. It significantly protects its innate structures from self-repression when nanC*&spot42(or nanC&*spot42 ) was assembled together.
Moreover, fortunate as it is that complementary domain between nanC and spot42 coincidently avoids that between galK and spot42, which prevent *spot42 from deprivation of inhibition of galK::GFP expression. The complementary pairing of two pairs are shown below.

The secondary structures of spot42 , nanC, *spot42::nanC and nanC*::spot42 is shown below.

Here we can see that the structure is highly compatible when nanC*::spot42 (or nanC::*spot42)assembled, involving previously described dsrA::spot42(or rpos::spot42). It further inspired us to carry out experiments to verify our designs.
Future workOur experiment of those two designs are still in progress. Actually all the artificial sRNA assemblies have already succeeded, though have not been transformed into E.coli with combined plamids carrying our reporter genes. Provided that the-last-run transformation is finished, we could see whether our designs will work well or not. It is exciting that we would check them before the Asian Jamboree and showcase our results of our brand-new ‘ratio sensor’
Artificial region assembly & Testing quantitatively
Assembly method
We amplify artificial region by PCR from DH5α,using primers with ClaI/SalI site;
ClaI site and SalI site are well-selected restriction sites,they are used to assembly the artificial region to the 5’ end of spot42,are compatible for all of the Ampr and Chlr vectors(no unexpected restction sites).
Testing method
All the sRNA generators with artificial region were tested with inducer gradient, to verify spot42 of different levels repression efficiency in vivo.(Results)
We tested Comparators ligated with srlA/ytfJ/nanC buffer RNA with aTc/IPTG 2-dimension gradient using well designed protocol,the data is given in results page,while the data of ratio sensor is still in progress.
Adjustment
We also made some adjustment on the mRNA synthesis rate(αm) by replacing our device to different copy number vectors.We assemblied two sRNA generator together to J61002(Ampr) while ligating GFPmRNA_Generator to pSB4C5(Chlr), to perform double-plasmids transformation.But the function of buffer RNA didn’t work well due to unclear reasons.

Note
1/ColE1 is a medium copy number replication origin(~70 copies per cell),whereas p15A is a low one(~5 copies per cell).
2/The efficiency of endogenous terminator of spot42 remains unclear. Another consideration of swiching vectors is to avoid the readthrough of spot42.
Some evidence suggests it’s not very strong: when induces spot42 in the upstream of GFP_Generator with very high aTc concentration, the GFP level soared to a very high level(even higher than GFP_generator alone), made it elusive on this paradox.
 "
"
 Home
Home
 HumanPractice index
HumanPractice index
 JudgingForm
JudgingForm
 Contact Us
Contact Us
 Project Overview
Project Overview
 Sensor
Sensor
 Decision-making Device
Decision-making Device
 Gas vesicle
Gas vesicle
 ODEModel
ODEModel
 Parameter Sensitivity Analysis
Parameter Sensitivity Analysis
 Parameter Sweep
Parameter Sweep
 Noise Analysis
Noise Analysis
 HumanPractice Overview
HumanPractice Overview
 Meeting and Academic Communication
Meeting and Academic Communication
 Camps, Class and Lectures
Camps, Class and Lectures
 Special HP
Special HP
 Team Members
Team Members
 Instructors
Instructors
 Acknowledgement&Cooperation
Acknowledgement&Cooperation
 Lab
Lab
 Parts
Parts
 Safety
Safety
 Labnote
Labnote
 Modeling Note
Modeling Note
 Protocols
Protocols



