Team:Bielefeld-Germany/Labjournal/week14
From 2012.igem.org
(Difference between revisions)
(→Week 14 (07/30 - 08/05/12)) |
(→weekly seminar) |
||
| Line 49: | Line 49: | ||
==Week 14 (07/30 - 08/05/12)== | ==Week 14 (07/30 - 08/05/12)== | ||
=== weekly seminar === | === weekly seminar === | ||
| - | + | * From now on, there will be two weekly meetings: monday, 12.15 and thursday 9.00. | |
| + | * Nadine sent all of us a poll, answers will be used on the wiki. | ||
| + | * For picture uploads we will use the name: Bielefeld2012_x. | ||
| + | * Which sewage treatment plant uses activated carbon in the third stage of treatment? | ||
| + | * On 25th of | ||
| + | |||
===Monday July 30th=== | ===Monday July 30th=== | ||
* '''Team Site Directed Mutagenesis:''' Plasmid-isolations of all the bpul- and xccl-mutants. | * '''Team Site Directed Mutagenesis:''' Plasmid-isolations of all the bpul- and xccl-mutants. | ||
Revision as of 14:37, 25 September 2012
Contents |
Week 14 (07/30 - 08/05/12)
weekly seminar
- From now on, there will be two weekly meetings: monday, 12.15 and thursday 9.00.
- Nadine sent all of us a poll, answers will be used on the wiki.
- For picture uploads we will use the name: Bielefeld2012_x.
- Which sewage treatment plant uses activated carbon in the third stage of treatment?
- On 25th of
Monday July 30th
- Team Site Directed Mutagenesis: Plasmid-isolations of all the bpul- and xccl-mutants.
- Team Cellulose Binding Domain:
- Redesigned the primer-sequences another time giving the CBDs a few additional amino acids from the by Protein-BLAST predicted domain. [http://partsregistry.org/Part:BBa_K863102 CBDcex]: 4 AS N-terminal, 2 C-terminal. [http://partsregistry.org/Part:BBa_K863112 CBDclos]: 2 N-terminal (starting with a natural ATG) and 2 AS C-terminal.
- Prepared <partinfo>BBa_K392014</partinfo> for Sequencing
- Cloning of Bacterial Laccases:
- The PCR products from the 23th July was digested with EcoRI and SpeI same as the pSB1C3 vector. Then the ligation was set and transformed into competent E. coli KRX cells.
- Since we haven't got any more PCR products we set a PCR. Only S. goettingen worked.
- Team Fungal and Plant Laccases: Digestion of tvel5 PCR product with prefix and suffix ends for cloning in pSB1C3 and digest of laccase with the overhangs for cloning in shuttle vector. Additionally we did the PCR on tvel35 with both primer pairs again. This time we lowered the annealing temperatures and got products with both primer pairs.
- Team Cultivation & Purification:
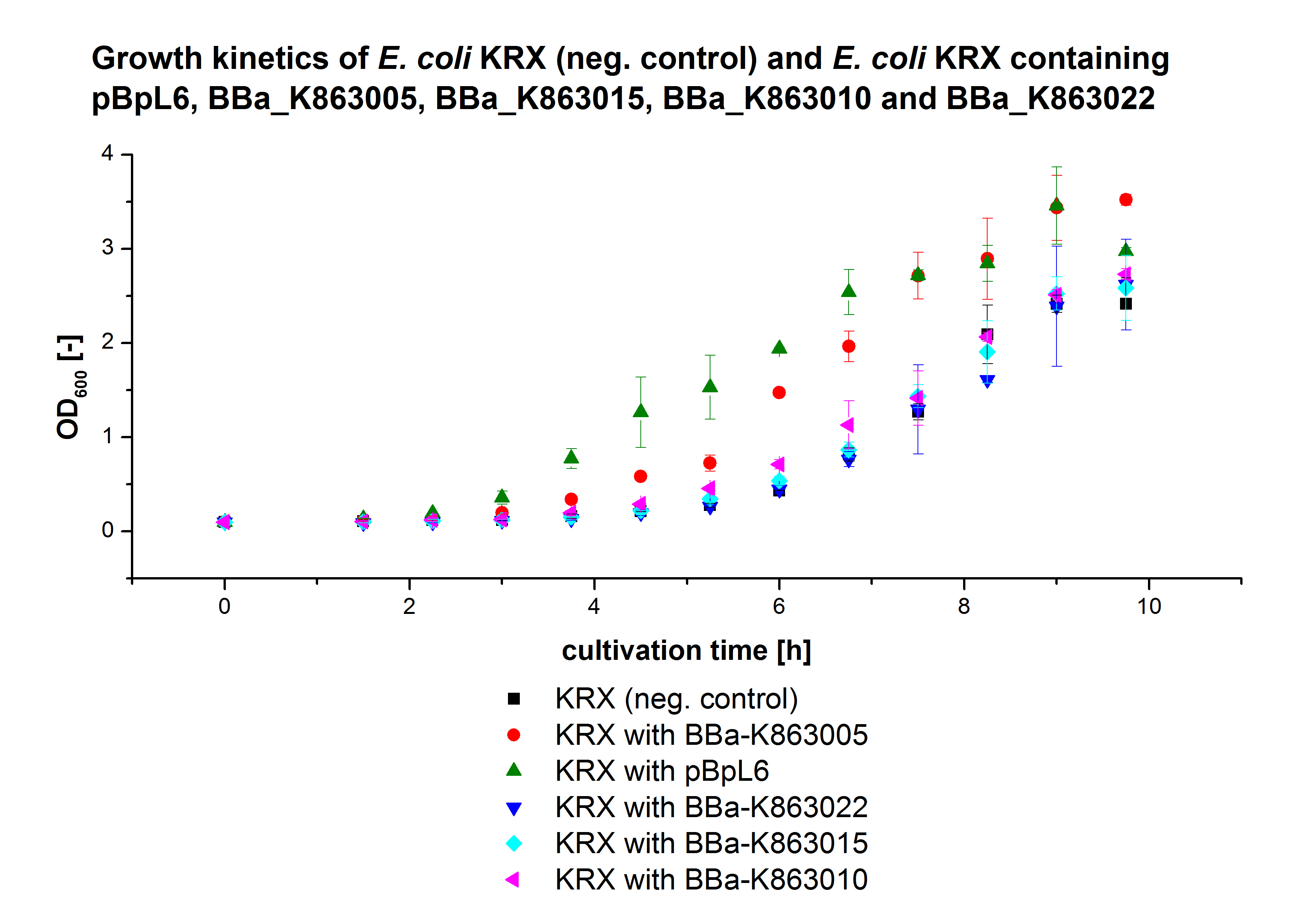
Figure X: Growth kinetics of cultivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and pBpL6. Parameters: autoinduction medium, 0,25 mM CuCl2, 28 °C.
- We made the SDS-Pages for the cultivation from 07/27, but they did not seem to be promising.
- We started another cultivation of E.coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and pBpL6.
- Settings: 300 mL flasks without baffles, final volume: 60 mL, autoinduction medium, 0,25 mM CuCl2, 28 °C.
- A growth kinetics was recorded every 45 minutes.
- We decided that we also need a positive control for the next cultivations, to see if our autoinduction medium works. We chose [http://partsregistry.org/Part:BBa_K525710 BBa_K525710]. Furthermore we will make cultivations in which we manually induce the expression after 4 hours.
- Made a preculture of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and pBpL6 as well as with [http://partsregistry.org/Part:BBa_K525710 BBa_K525710]. We used E. coli KRX as negative control.
Tuesday July 31st
- Team Cloning of Bacterial Laccases:
- After our laccases are not produced or not detectable we decided to try to express the laccases with a constitutive promoter. Therefore we searched for appropropriate promoters and found different promoters from the [http://partsregistry.org/Ribosome_Binding_Sites/Prokaryotic/Constitutive/Anderson Anderson promoter family]. We picked three different promoters with different promoter strengths. We chose the parts [http://partsregistry.org/wiki/index.php/Part:BBa_J23103 BBa_J23103], [http://partsregistry.org/wiki/index.php/Part:BBa_J23103 BBa_J23110] and [http://partsregistry.org/wiki/index.php/Part:BBa_J23103 BBa_J23117] and for all three promoters the RBS [http://partsregistry.org/wiki/index.php/Part:BBa_J23103 BBa_B0034]. The primers were designed that the FW and the RV primers anneal together to a short oligonucleotide with overhanging restriction site ends for EcoRI and SpeI. So we don’t have to cut the annealed primers, because the sites should appear with correct annealing of FW and RV primers. The goal is to clone the different promoters, the PCR products with the different laccase genes with His-Tag in pSB1C3 in one ligation step. Furthermore we want to exclude the possibility that the His-tag is the reason for no activity, so we also want to clone the gene sequences without a His-tag under control of a constitutive promoter in pSB1C3.
- Additionally we want to produce the constructs with a new T7 promoter. After our laccases were not expressed we now think that maybe the RBS <partinfo>BBa_B0034</partinfo>, which we changed in our primers from originally 5' aAagaggagaaa 3' to 5' aGagaggagaaa 3' is not or poorly recognized from the ribosomes in the cells.
- We have a primer pair from last year with the T7 promoter sequence with the RBS BBa_B0034. The primers can be annealed to an oligonucleotide. After boiling the primers and cooling down there should be an oligonucleotide with an EcoRI and a PstI restriction site. We now want to assemble the promoter the laccase genes and the pSB1C3 vector in one step. The pSB1C3 backbone was digested with EcoRI and PstI, the laccases with XbaI and PstI, and the promoter has the restriction sites SpeI and EcoRI (we have to digest the promoter with SpeI, because the original restriction site is PstI).
- Team Fungal and Plant Laccases:
- Purification of tvel35 PCR product and digestion for cloning in pSB1C3 backbone.
- Ligation of tvel5 laccase with pSB1C3 backbone.
- Team Site Directed Mutagenesis: Plated four colonies per dish of tvel-t243g and tvel-t1161a for plasmid-isolation.
- Test-restriction of the xccl-mutants showed that no colony was mutated correctly
- The second site directed mutagenesis of bpul-g2317t could not be rated by test-digestion, since the mutation at 2317 is not because of a illegal restriction site, but a amino acid alternating mutation.
- Made four bpul-colonies (with both mutations) ready for sequencing
- plated a four more colonies of xccl (both SDM-sites) for plasmid-isolation
- Team Cultivation & Purification:
- Made SDS-Pages of cultivation from 07/30.
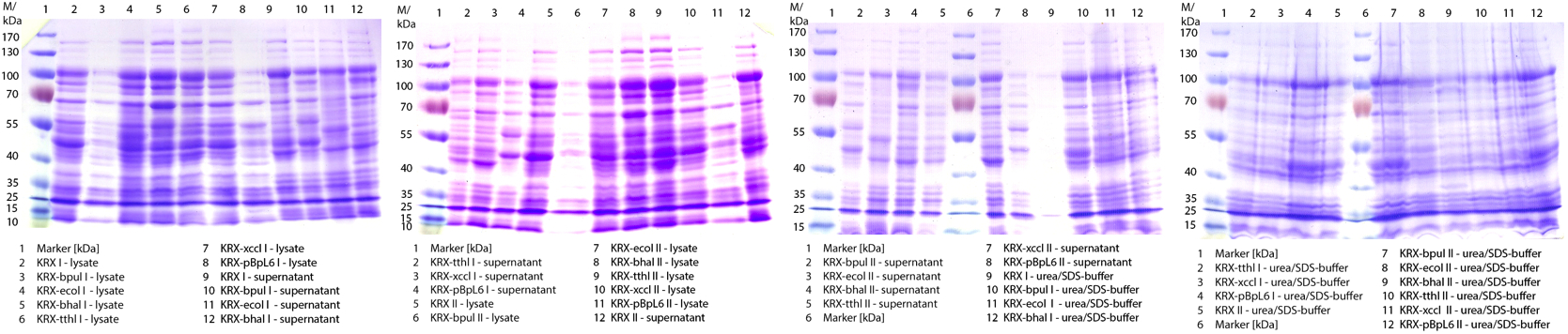 Figure 1: Cultivation from 07/30 of E.coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and pBpL6. 60 mL in 300 mL flasks without baffles with autoinduction medium, 0,25 mM CuCl2, 28°C
Figure 1: Cultivation from 07/30 of E.coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and pBpL6. 60 mL in 300 mL flasks without baffles with autoinduction medium, 0,25 mM CuCl2, 28°C
- Made SDS-Pages of cultivation from 07/30.
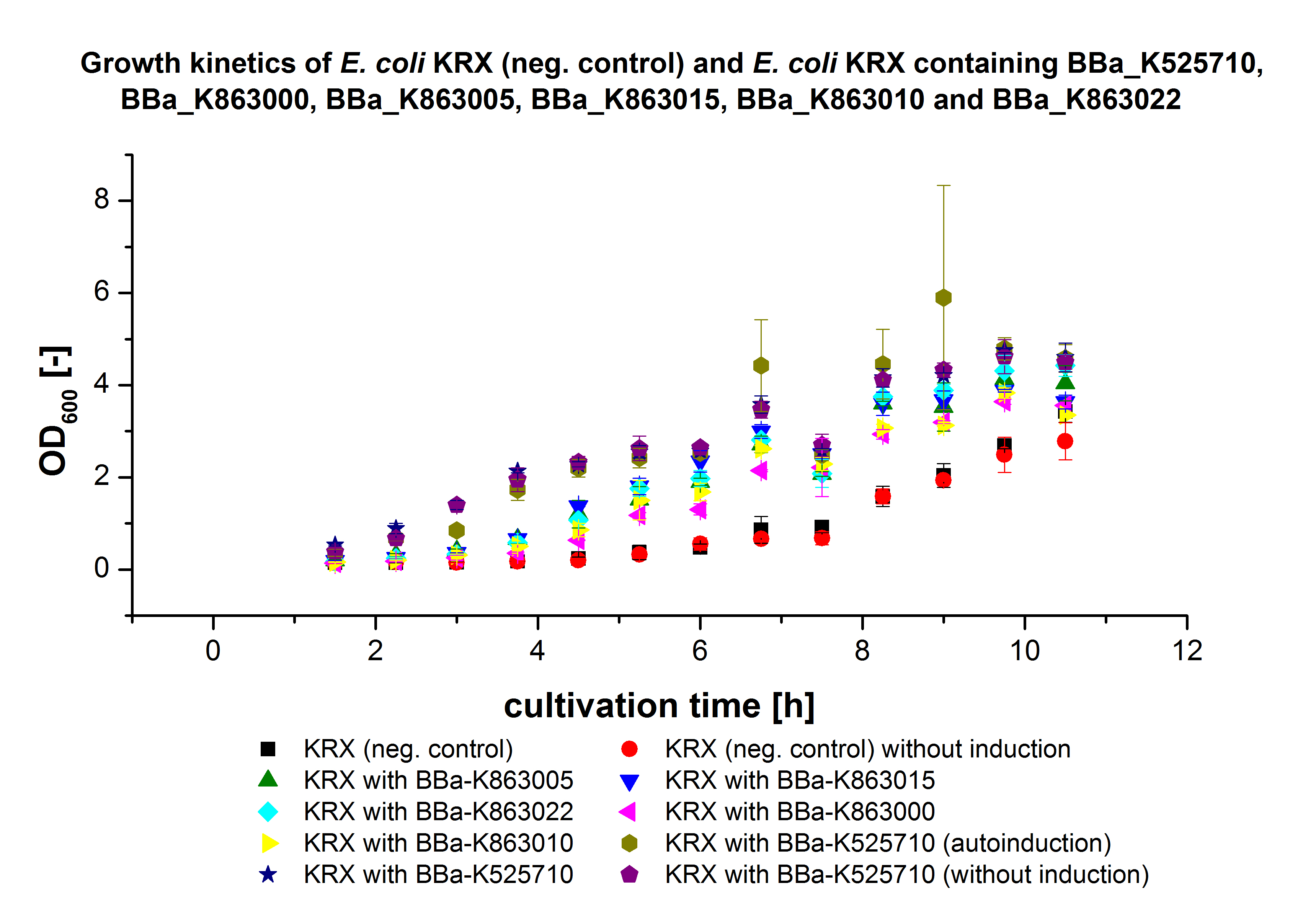
Figure X: Growth kinetics of cultivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and pBpL6. Parameters: autoinduction or manual induction, 0,25 mM CuCl2, 30 °C.
- Cultivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and pBpL6 as well as with [http://partsregistry.org/Part:BBa_K525710 BBa_K525710]. We used E. coli KRX as negative control.
- general settings: 300 mL flasks without baffles, final volume: 60 mL, 30 °C
- special settings:
- Cultivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and pBpL6 as well as with [http://partsregistry.org/Part:BBa_K525710 BBa_K525710]. We used E. coli KRX as negative control.
- a) E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/Part:BBa_K525710 BBa_K525710], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and pBpL6 were cultivated in LB media and manually induced after 4 hours with 0,1 % rhamnose.
- b) E. coli KRX without plasmid was cultivated with LB medium and induced after 4 hours with 0,1 % rhamnose.
- c) E. coli KRX with [http://partsregistry.org/Part:BBa_K525710 BBa_K525710] was cultivated equally to b)
- d) E. coli KRX without plasmid was cultivated with autoinduction medium and 20 µg/mL chloramphenicol as well as 100 µg/mL ampicillin as a control.
Wednesday August 1st
- Team Site Directed Mutagenesis: Plasmid-isolation of tvel- and the xccl-colony-dishes.
- Test-restriction with showed correct bands for two tvel and one xccl-colony.
- Made the correct xccl-plasmid ready for sequencing.
- Team Cloning of Bacterial Laccases:
- With the new T7 promoter we started new assemblies. We first want to try the new promoter with ecol. So wedigested our laccase PCR products from ecol with and without HIS tag for suffix insertion with XbaI and PstI, boiled and cooled down the joined T7 primer pairs and digested pSB1C3 backbone with EcoRI and PstI. If the plan works, the parts anneal to our final plasmid. We did the ligation and transformation of the approach into competent E. coli KRX cells.
- Team Cultivation & Purification:
- Harvesting and cell disruption via sonification of yesterday's cultivation.
Thursday August 2nd
- Team Wiki: This morning we met for taking a new picture of our group and individual pictures of everyone. Check out our beautiful team members here.
- Team Site Directed Mutagenesis: pfu-PCRs; two with the positive tvel-t243g-plasmids as templates and the tvel-t1161a primer-mix and another pfu-PCR with the positve xccl-g2247c-plasmid and the xccl-g3633c primer-mix
- Team Cloning of Bacterial Laccases:
- There were just red colonies on the plates from the transformation the day before. So we didn't do colony PCR and have to do the assembly again.
- Our Laccases genes from S. goettingen, S. tuebingen and S. roseochromogenes are around 2kb. The NEB-Phusion has a range of 1kb/30sec so we thought if we set the elongation time higher we might have products for S. tuebingen,S. roseochromogene so we did PCR. But the results showed that it doesn't depend on the elongation time from the Phusion because we haven't had any products.
- Team Fungal and Plant Laccases: Ligation of tvel5 and pcil35 in pSB1C3.
- Team Cultivation & Purification:
- Made SDS-Pages of cultivation from 07/31.

Figure 1: Cultivation from 07/31 of E.coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and pBpL6 as well as with [http://partsregistry.org/Part:BBa_K525710 BBa_K525710] (positive control) and E.coli KRX (negative control). 60 mL in 300 mL flasks without baffles, 30 °C. mI = manual induction, -I = without induction, auto-I = auto-induction.
Friday August 3rd
- Team Site Directed Mutagenesis:
- DpnI-digestion of the pfu-PCR-products (from yesterday). Transformed tevl- and xccl-double-mutants into XL1 Blue and plated them on selection-agar.
- Sequencing results:
- Both tthl-g2796a-mutations were successful
- ecol-g2307a has additional mutations
- Team Cellulose Binding Domain: Transformed the plasmids p714 (with the CDBcex domain) and p570 (with the CBDclos domain) we got from the fermentation group of Bielefeld University in KRX an plated them on kanamycin-selection-agar.
- Team Fungal and Plant Laccases: Transformation of tvel5 and tvel35 in pSB1C3 backbone.
- Team Cloning of Bacterial Laccases:
- Digest of ecol PCR products with and without HIS-Tag and ligation with pT7 in pSB1C3. Transformation of this ligations and transformation of [http://partsregistry.org/Part:BBa_K525710 BBa_K525710] for Team Cultivation.
- We did an annealing temp. gradient PCR with the Streptomyces bacteria. But expect 'S.goettingen neighter S. tuebingen not S. rosechromogenes did not work.
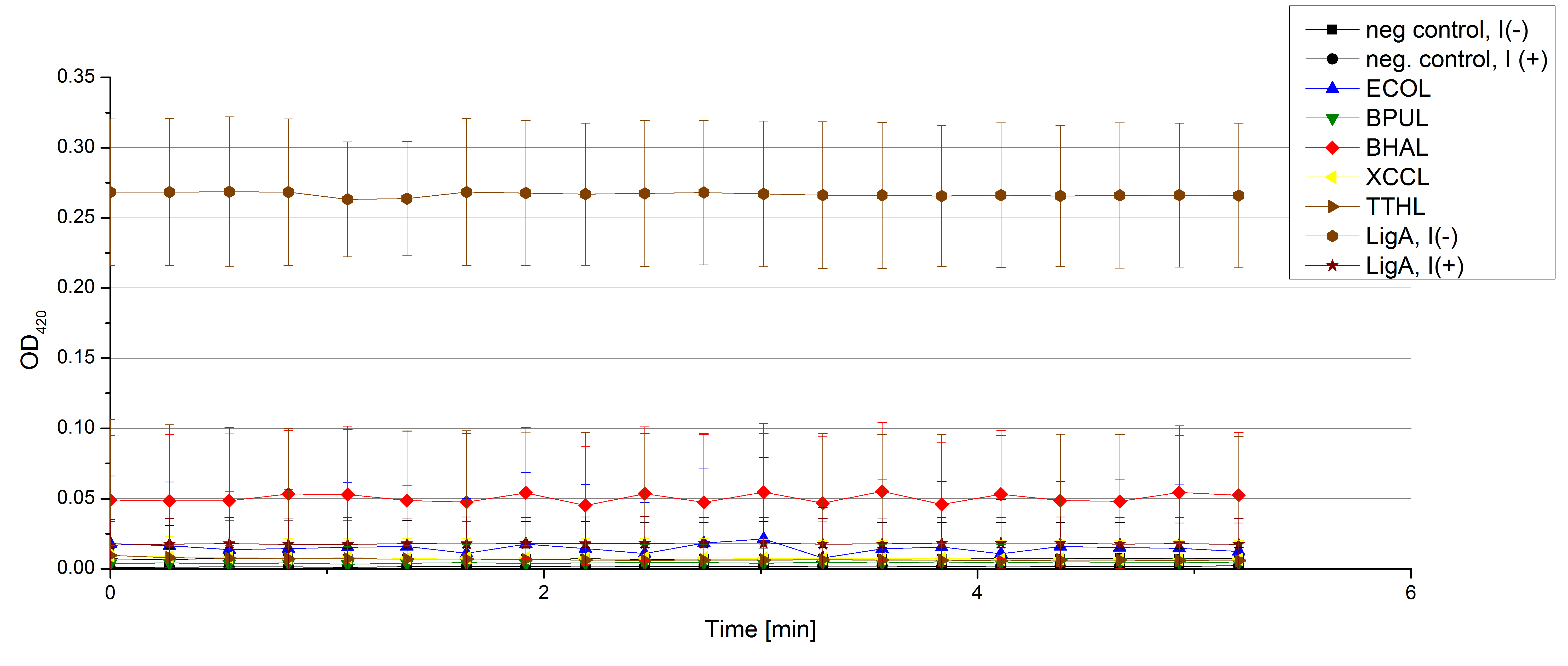
- Team Activity Tests: Our lovely Team Cultivation started new cultivations on 07/30 and we got the products. More precisely we got the supernatant of the lysis Team Cultivation did before. We tested 198 µL of each sample with 2 µL ABTS for the activity reaction, but sadly we couldn't detect anything (see Fig. 1).
Saturday August 4th
- Team Site Directed Mutagenesis: Plated four double-mutant-colonies from each transformation (both tvel-mutants and the xccl-mutant on selection-agar for plasmid isolation.
- Team Cellulose Binding Domain:
- The transformation of p570 brought up only a few colonies, so we took a few and plated them again on a selection-agar-dish.
- Plasmid-isolation of p714
- Team Cloning of Bacterial Laccases:
- We did the transformation of the ligations from August 1st again.
- Team Cultivation & Purification:
- We prepared precultures of E.coli KRX without plasmid and E.coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], pBpL6 or [http://partsregistry.org/Part:BBa_K525710 BBa_K525710] as positive control.
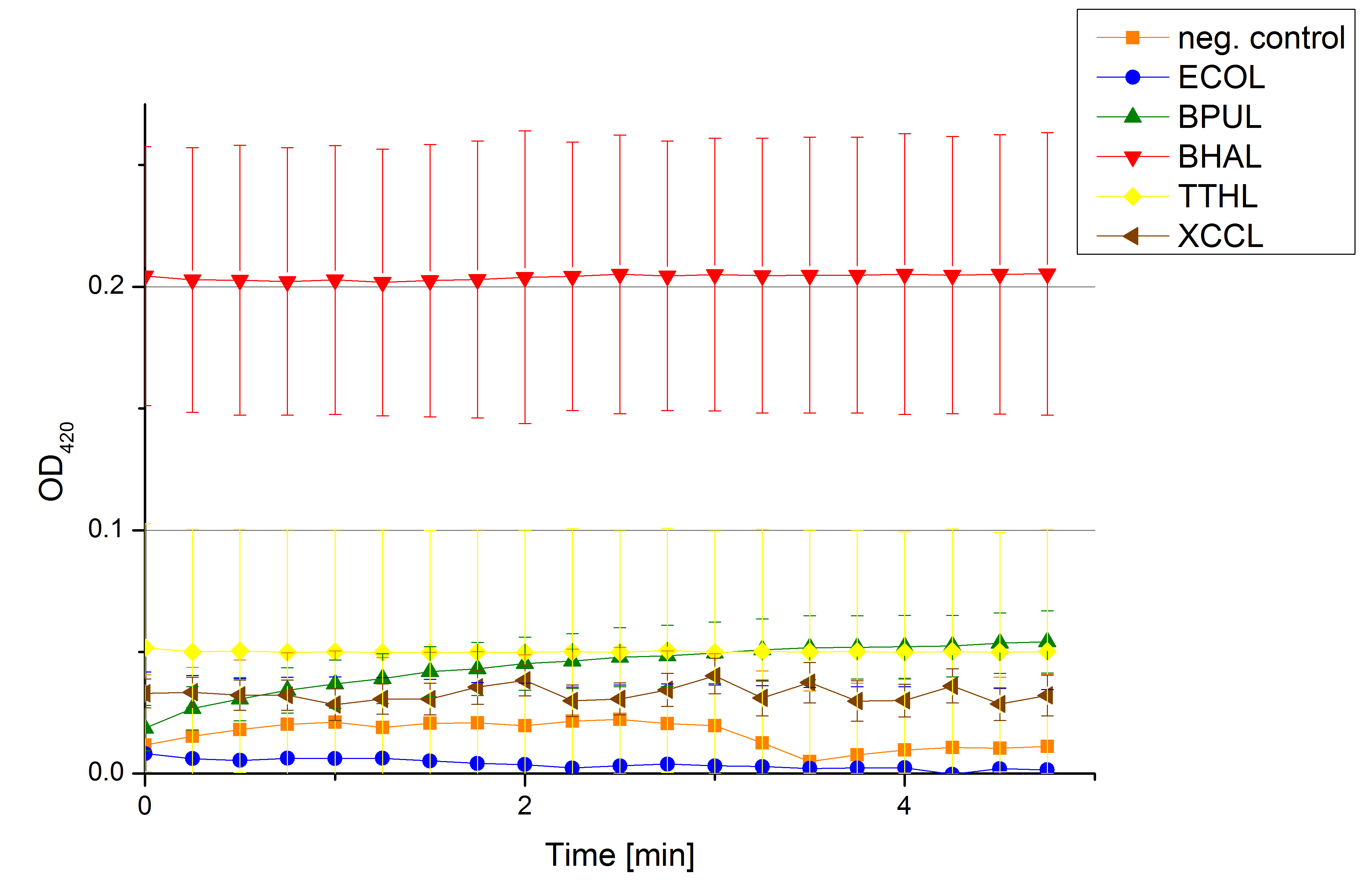
Check for active laccases in supernatants from lysed samples of the last cultivation ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005/ ECOL],[http://partsregistry.org/wiki/index.php/Part:BBa_K863000/ BPUL],[http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020/ BHAL],[http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010/ TTHL] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015/ XCCL]) (n=4).
- Team Activity Tests: Another cultivation ended and we are happy to measure the supernatants from the 31st September cultivated samples. We used 158 µL of the supernatant with 40 µL and added 2 µL of 20 mM ABTS to measure some activity (Fig. 1). Nevertheless we couldn't see any increase in absorbency at 420 nm after 5 minutes, indicating, that we have no active laccase in the supernatants.
Sunday August 5th
- Team Site Directed Mutagenesis: Plasmid-isolation of the xccl- and tvel-double-mutants.
- Test-restriction showed two positive plasmids on the xccl-g2247c and xccl-g3633c-mutagensis and no positve tvel.
- plated two colonies of the ecol-g2307a-mutagenesis and plated them on select-agar for plasmid-isolation.
- Prepared the two positive double mutants of the xccl-plasmid for sequencing.
- Team Cellulose Binding Domain:
- Transformated BBa_I13522 in KRX and plating it on AMP-selection-agar.
- Plasmid-isolation of p570
- PCR of [http://partsregistry.org/Part:BBa_K863103 CBDcex] (417 bp) and [http://partsregistry.org/Part:BBa_K863113 CBDclos] (369 bp) followed by test-gel-electrophoresis showed bands of the correct size and PCR-Clean-up
- Restriction of PCR-products and pSB1C3_RFP with XbaI and PstI.
- Team Cloning of Bacterial Laccases:
- The assemblies don't work by now in one step by now, we always get just red colonies from the original RFP plasmid. So we started to clone one part after another in pSb1C3 backbone. For this reason we want to ligate the ecol gene without any promoter in pSB1C3 backbone and in the next step do a prefix insertion with the promoter fragment. Therefore we did the restriction of ecol and ecol_HIS PCR products, the ligation in pSB1C3 and the transformation.
- Team Cultivation & Purification:
- We made another flask cultivation of E. coli KRX without plasmid and with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], pBpL6 and [http://partsregistry.org/Part:BBa_K525710 BBa_K525710] as positive control.
- general settings: 250 mL flasks without baffles, final volume: 50 mL, LB medium
- additional settings:
- We made another flask cultivation of E. coli KRX without plasmid and with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], pBpL6 and [http://partsregistry.org/Part:BBa_K525710 BBa_K525710] as positive control.
- a) 2 flasks of each culture were inducted with 0,1 % rhamnose after 4 hours of cultivation
- b) 2 flasks of each culture were not induced.
- We recorded the growth kinetics once per hour.
| 55px | | | | | | | | | | |
 "
"