Team:Bielefeld-Germany/Labjournal
From 2012.igem.org
JSchirmacher (Talk | contribs) (→Friday September 21st) |
(→Wednesday August 15th) |
||
| Line 2,029: | Line 2,029: | ||
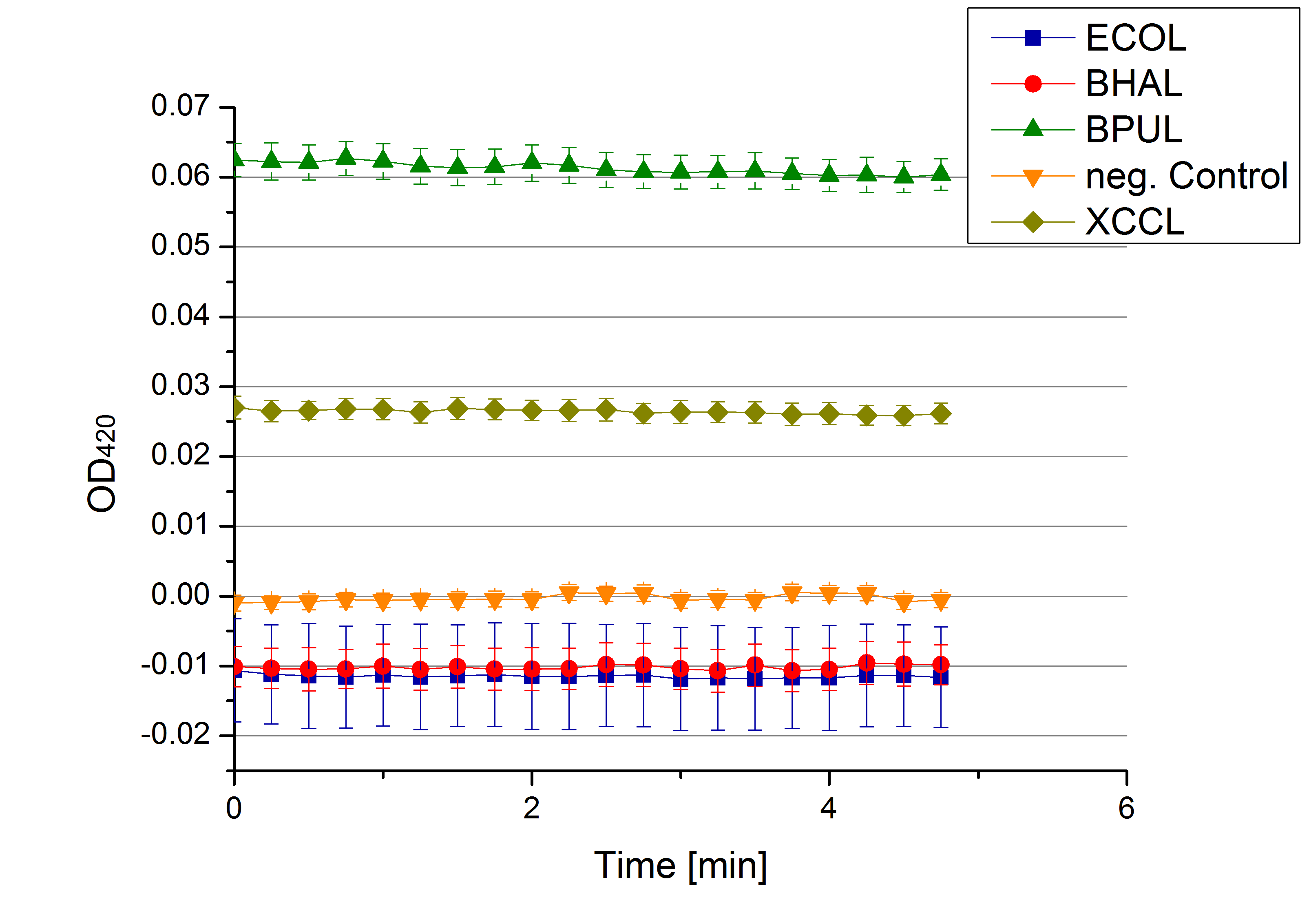
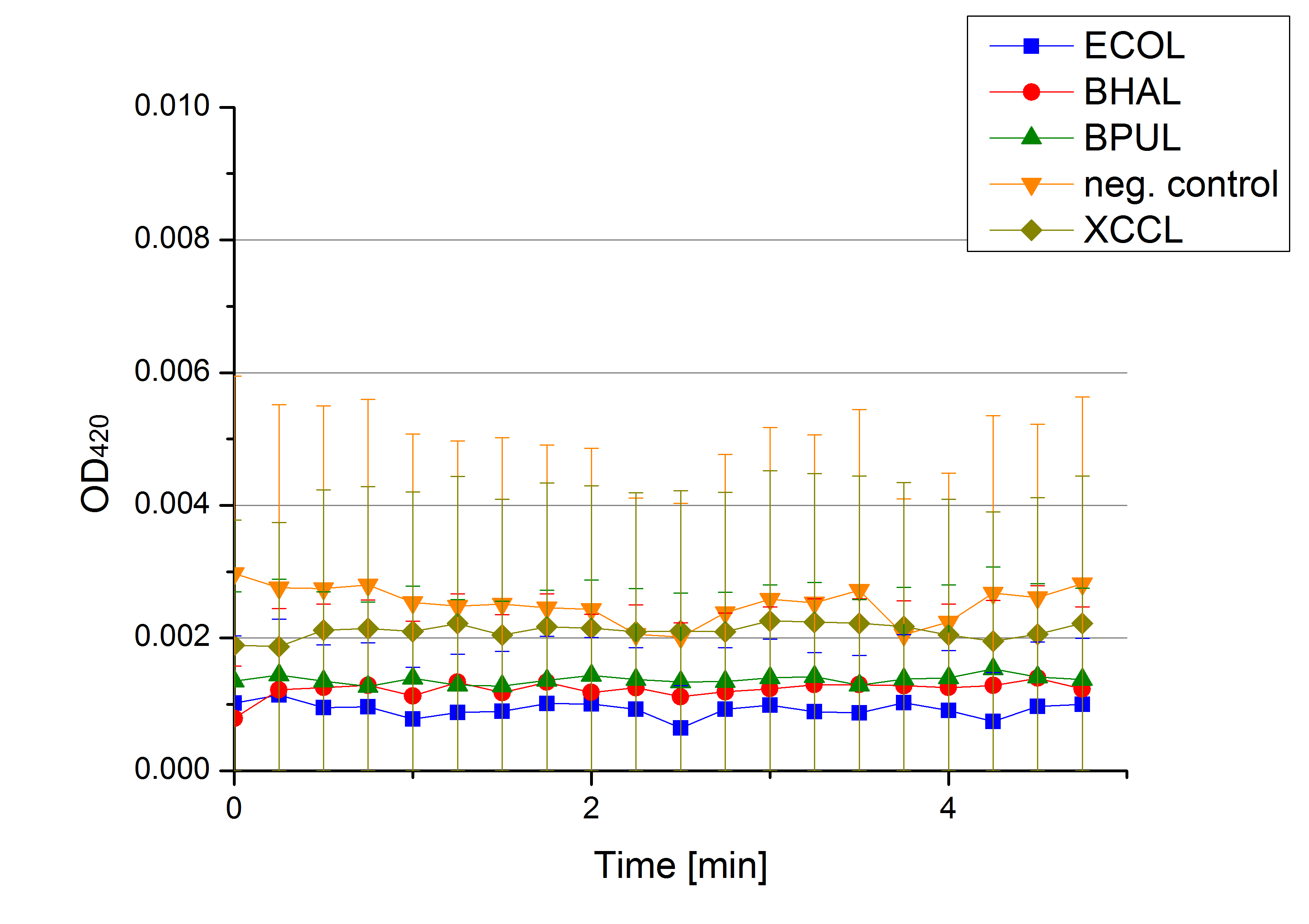
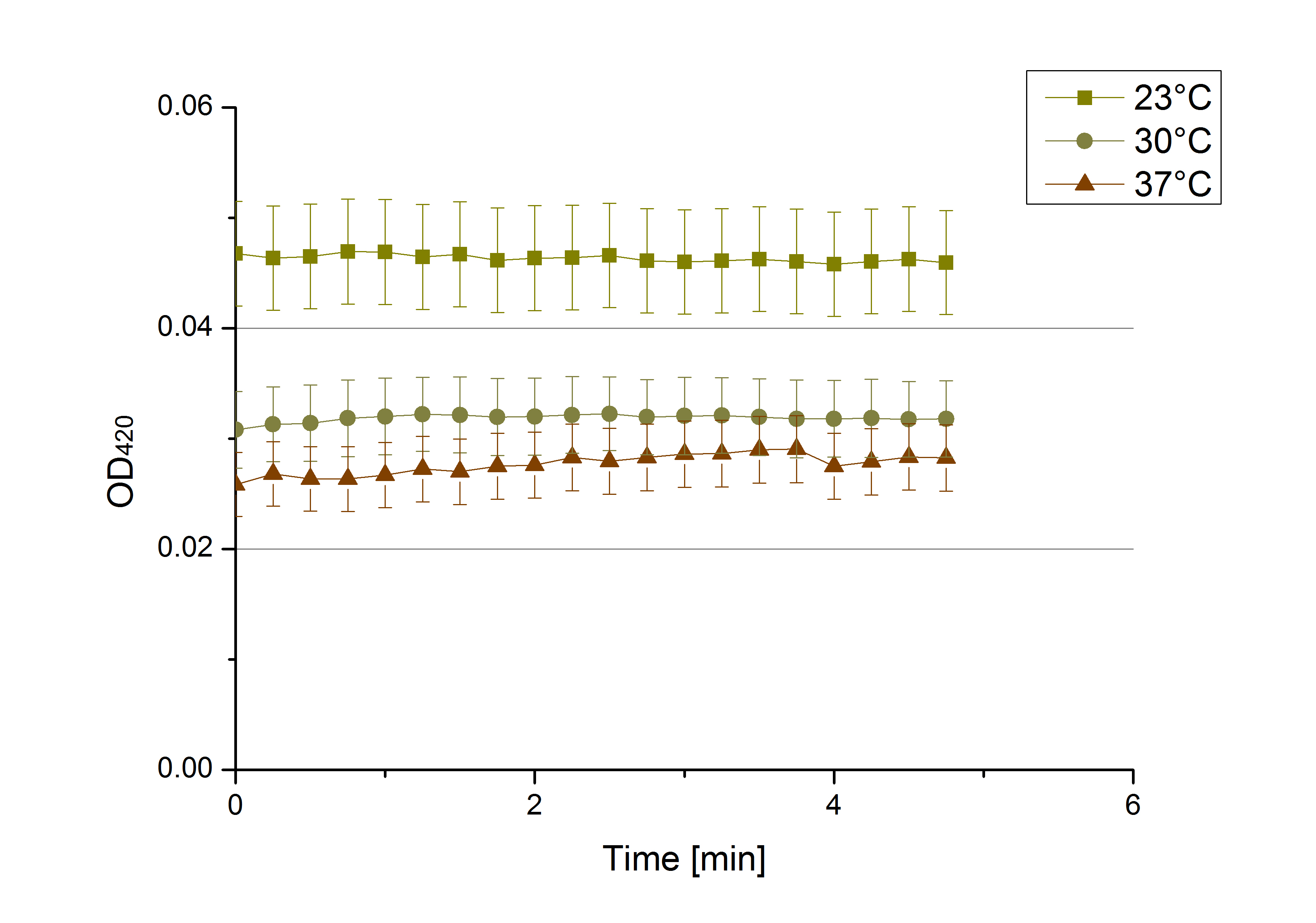
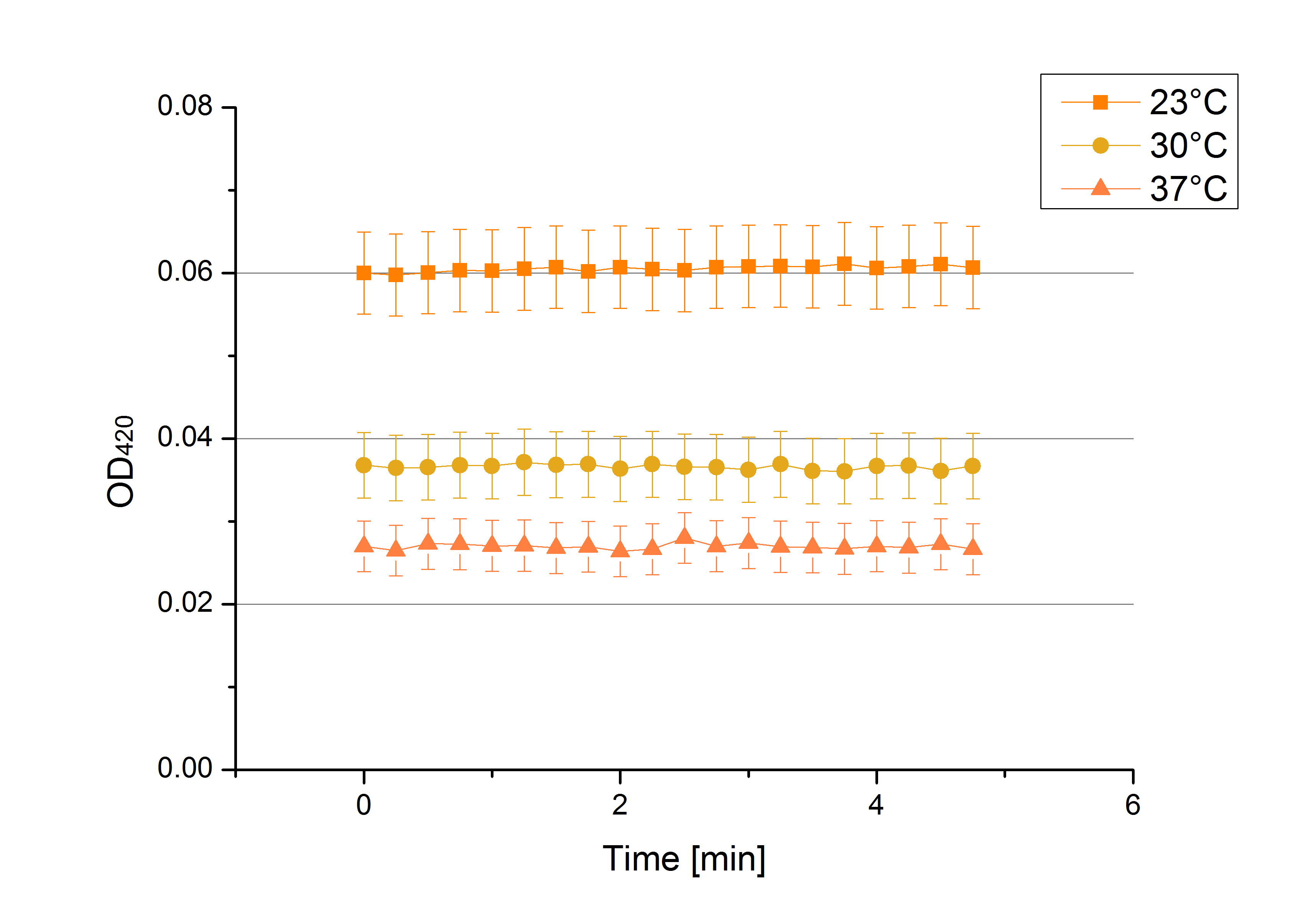
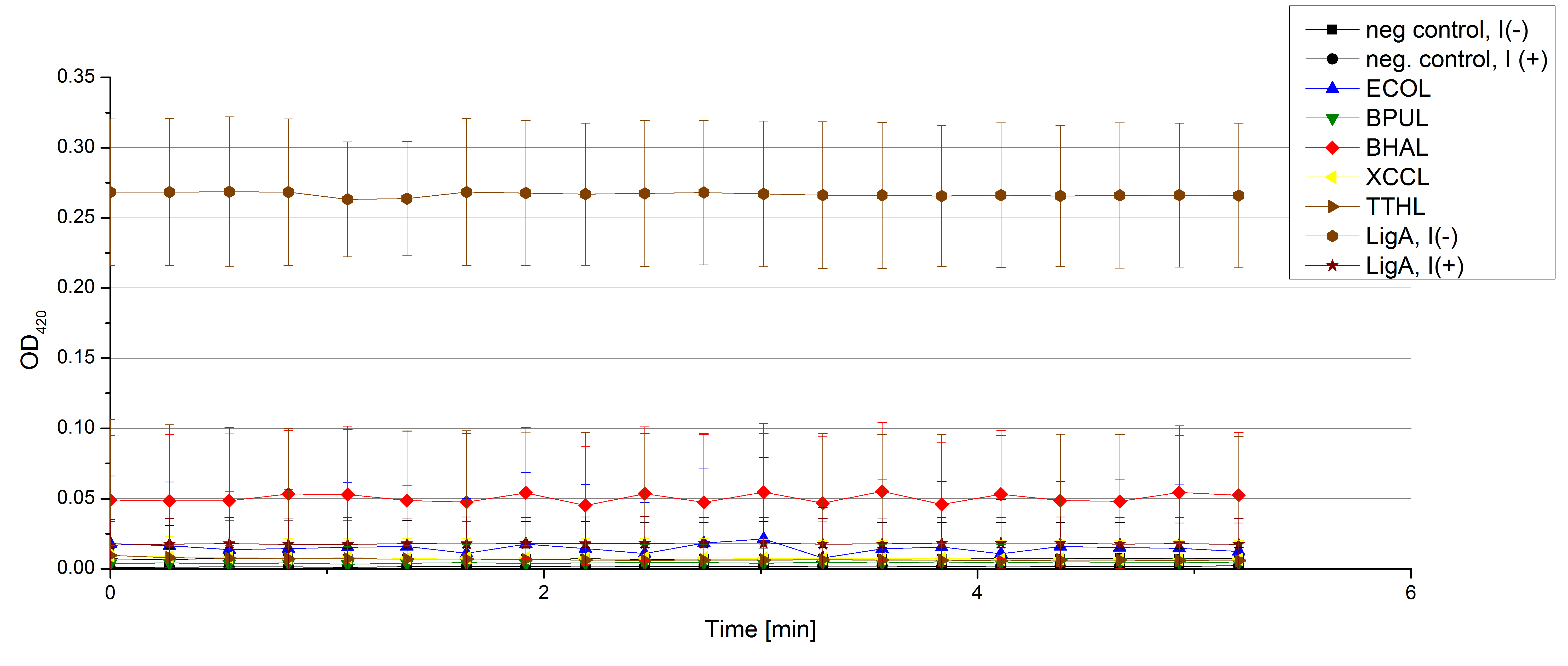
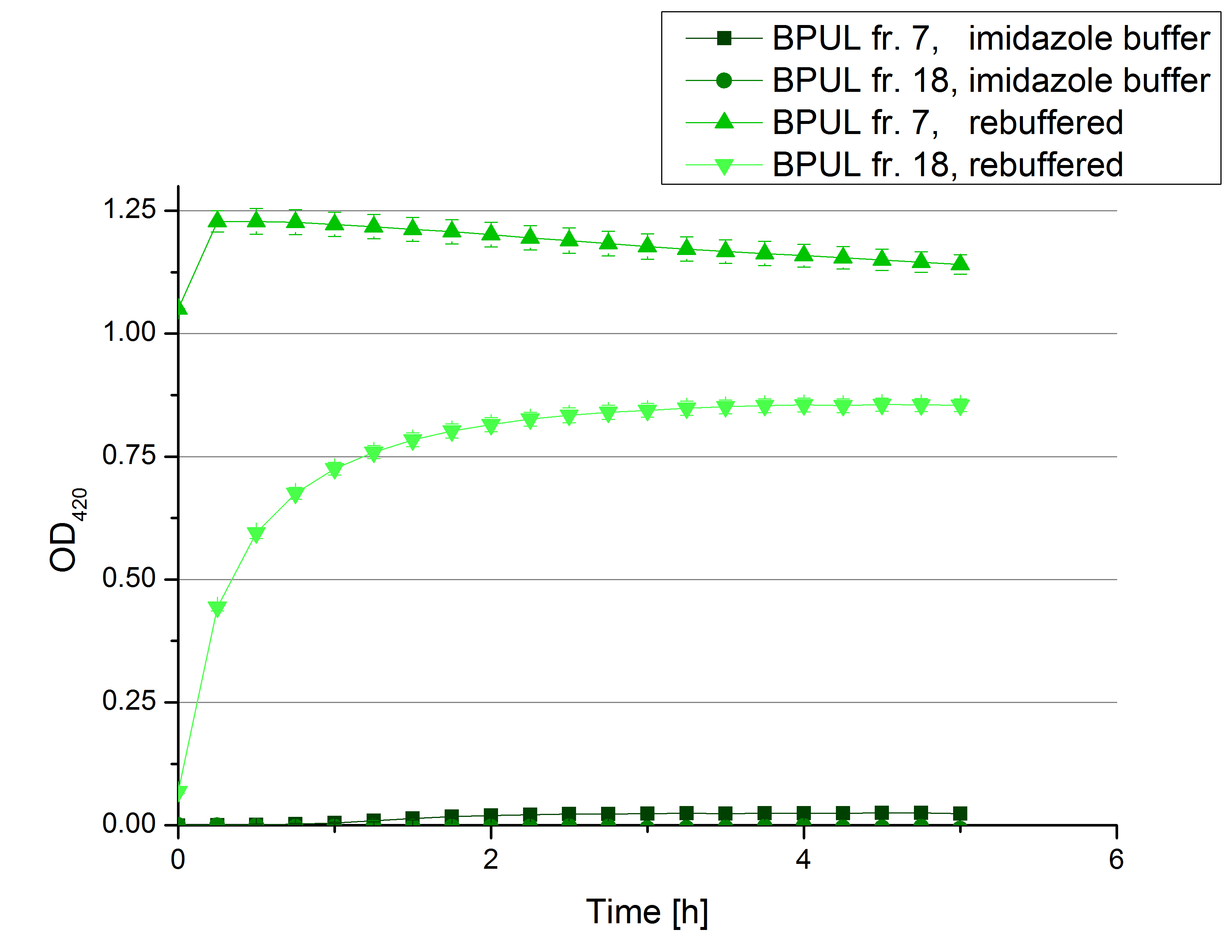
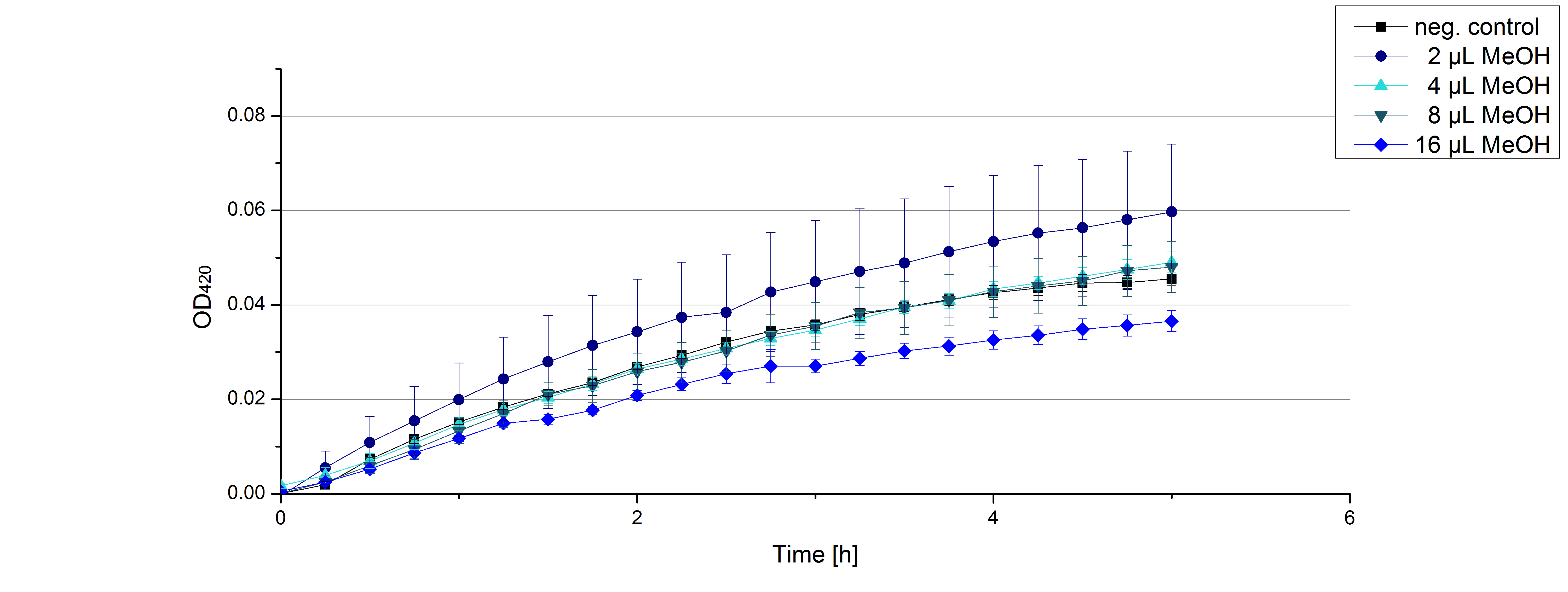
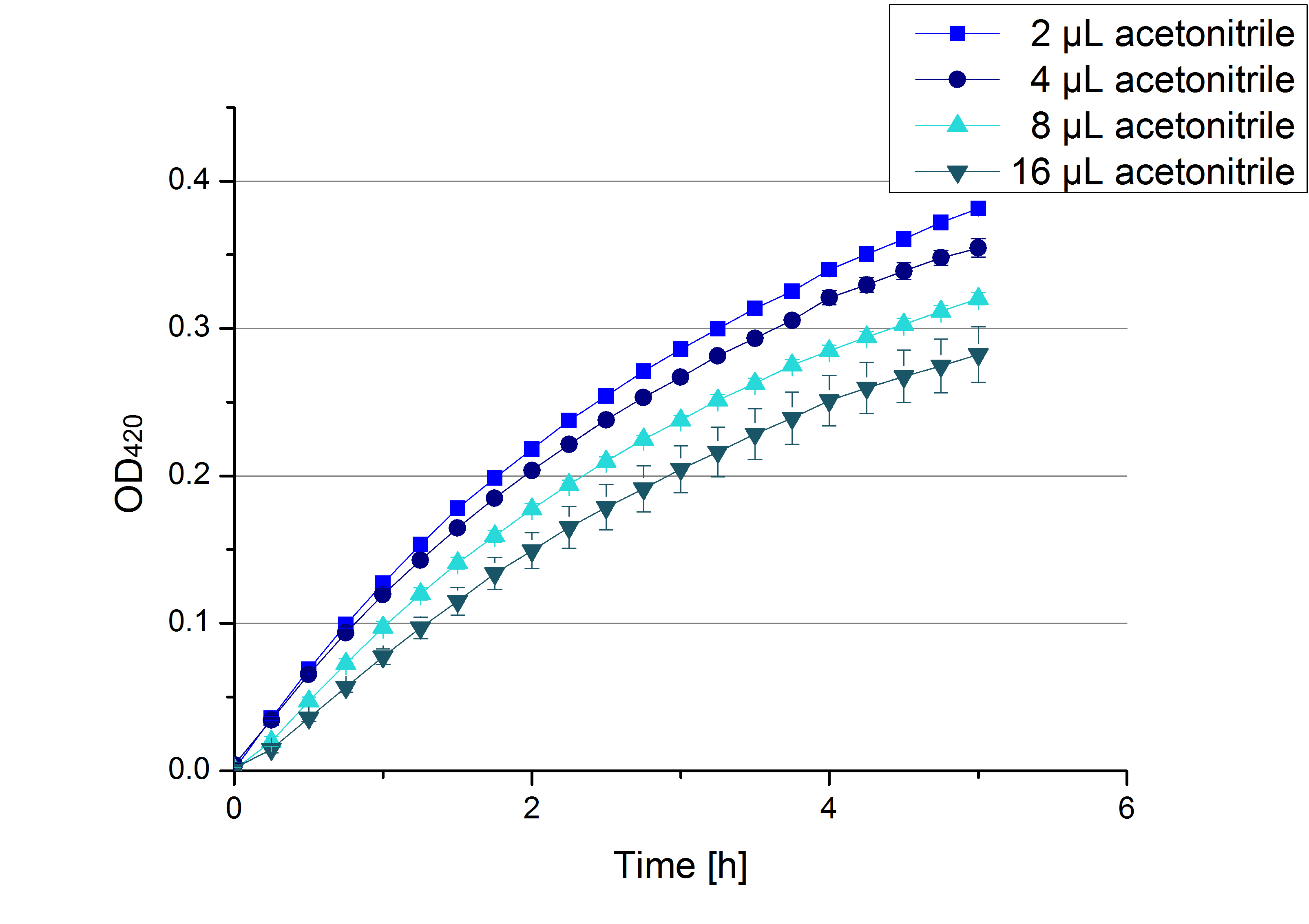
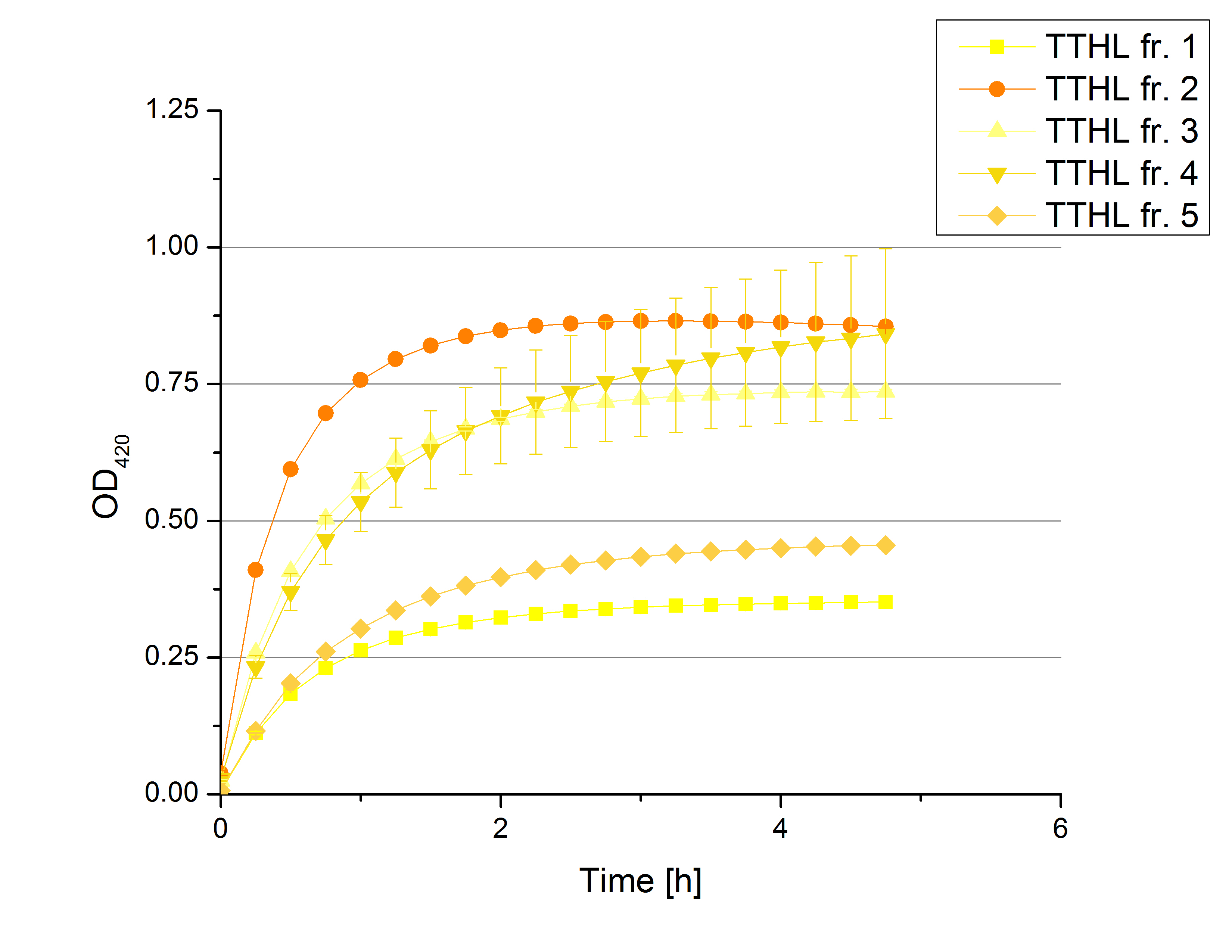
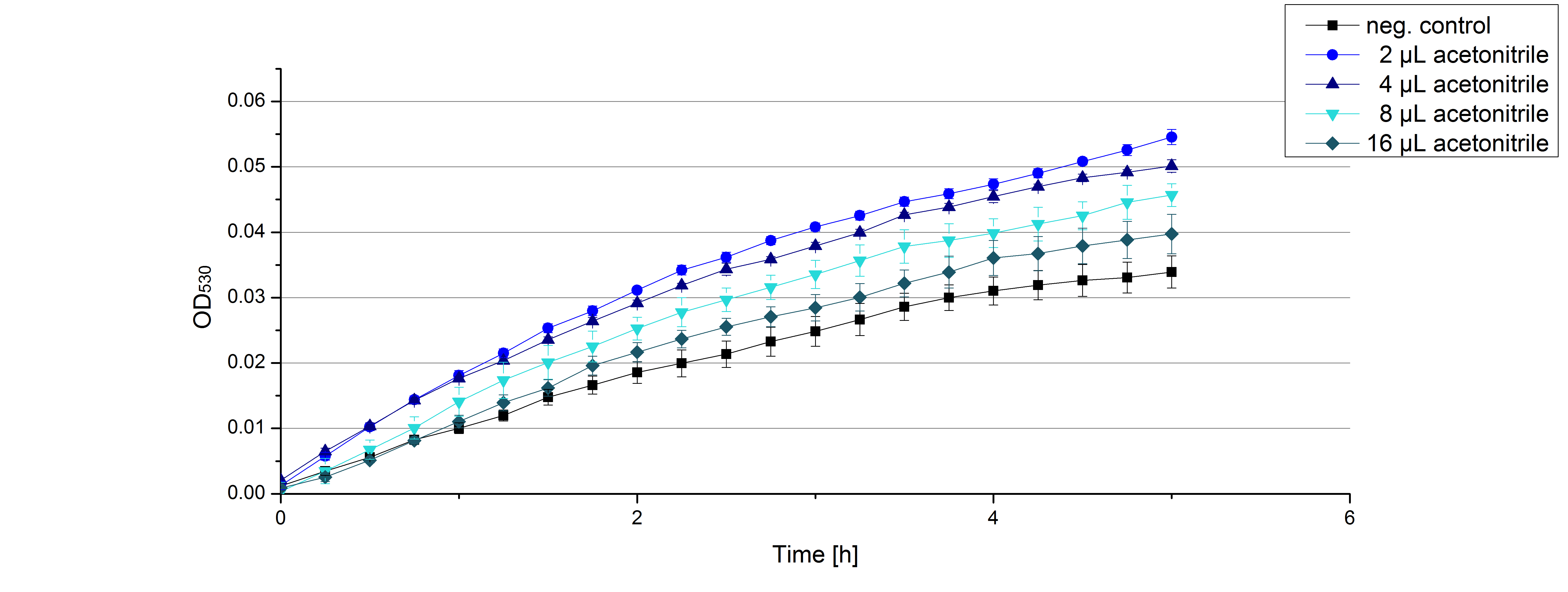
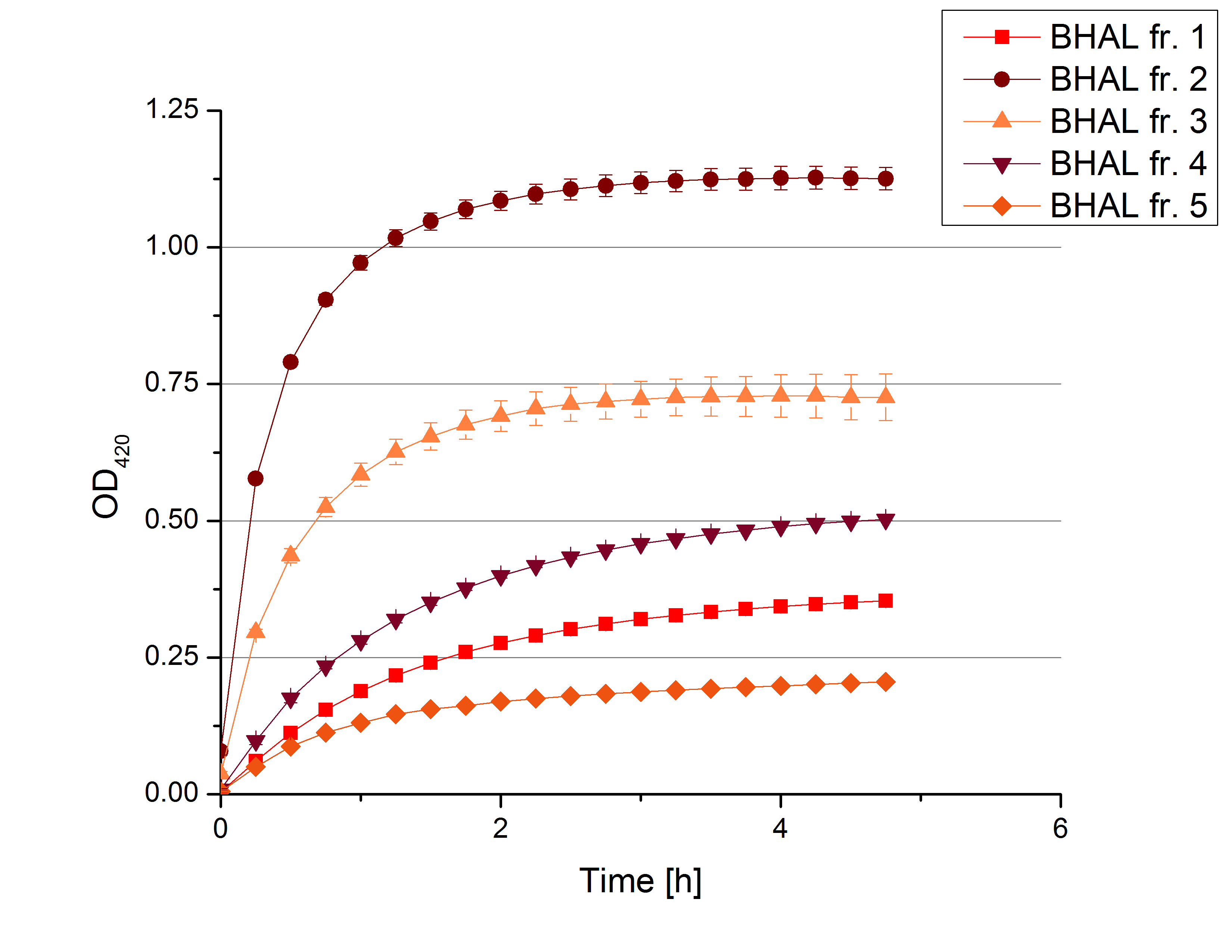
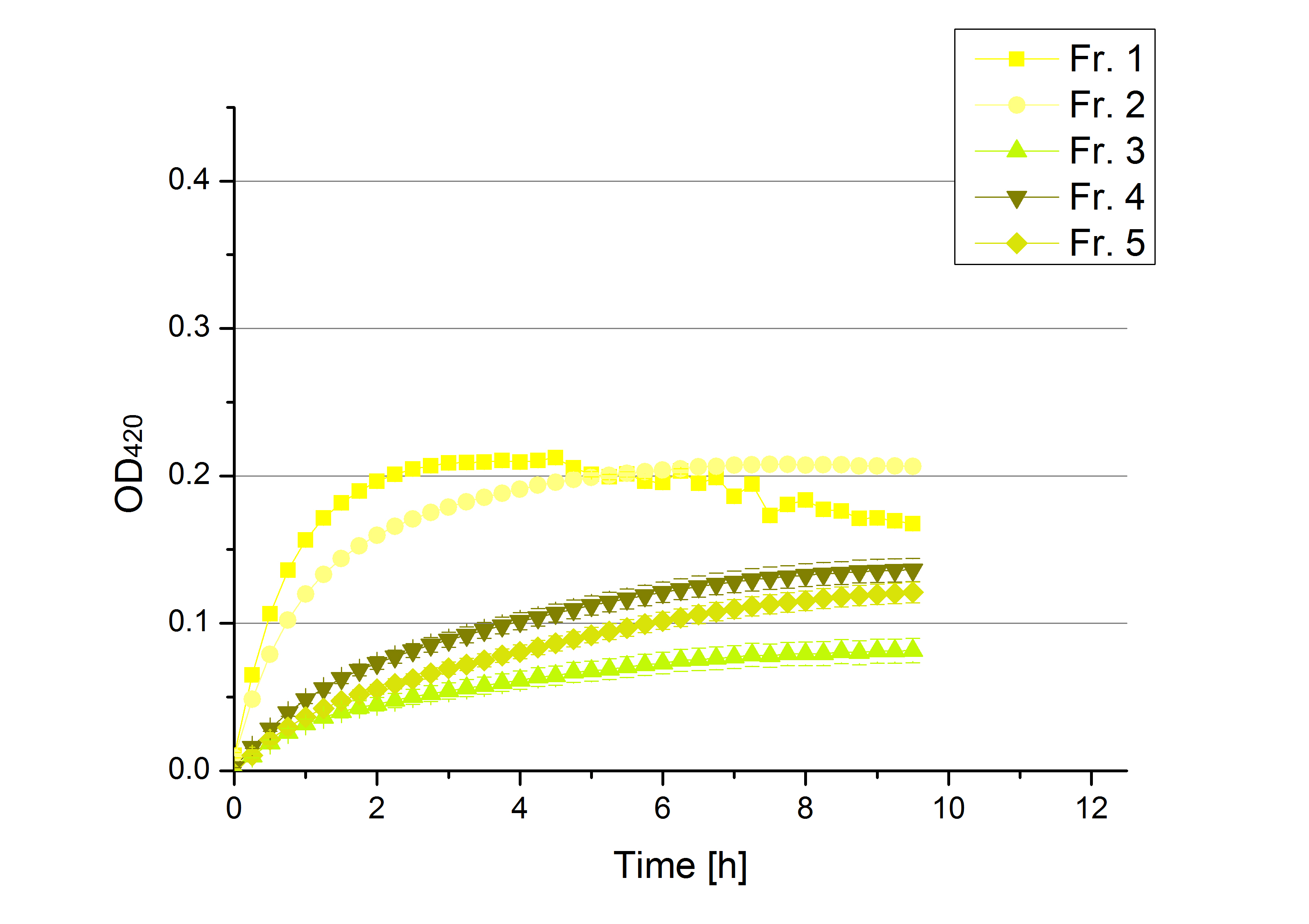
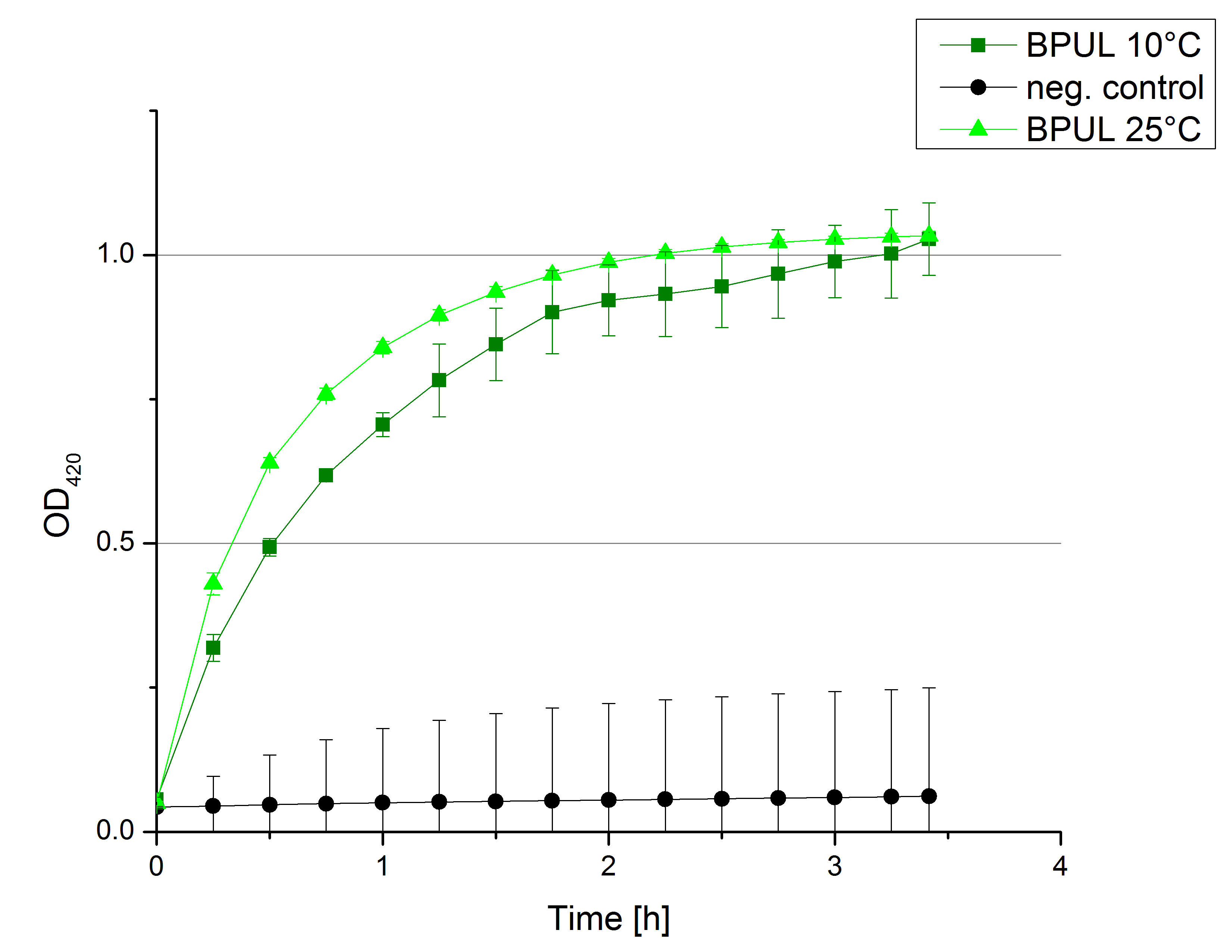
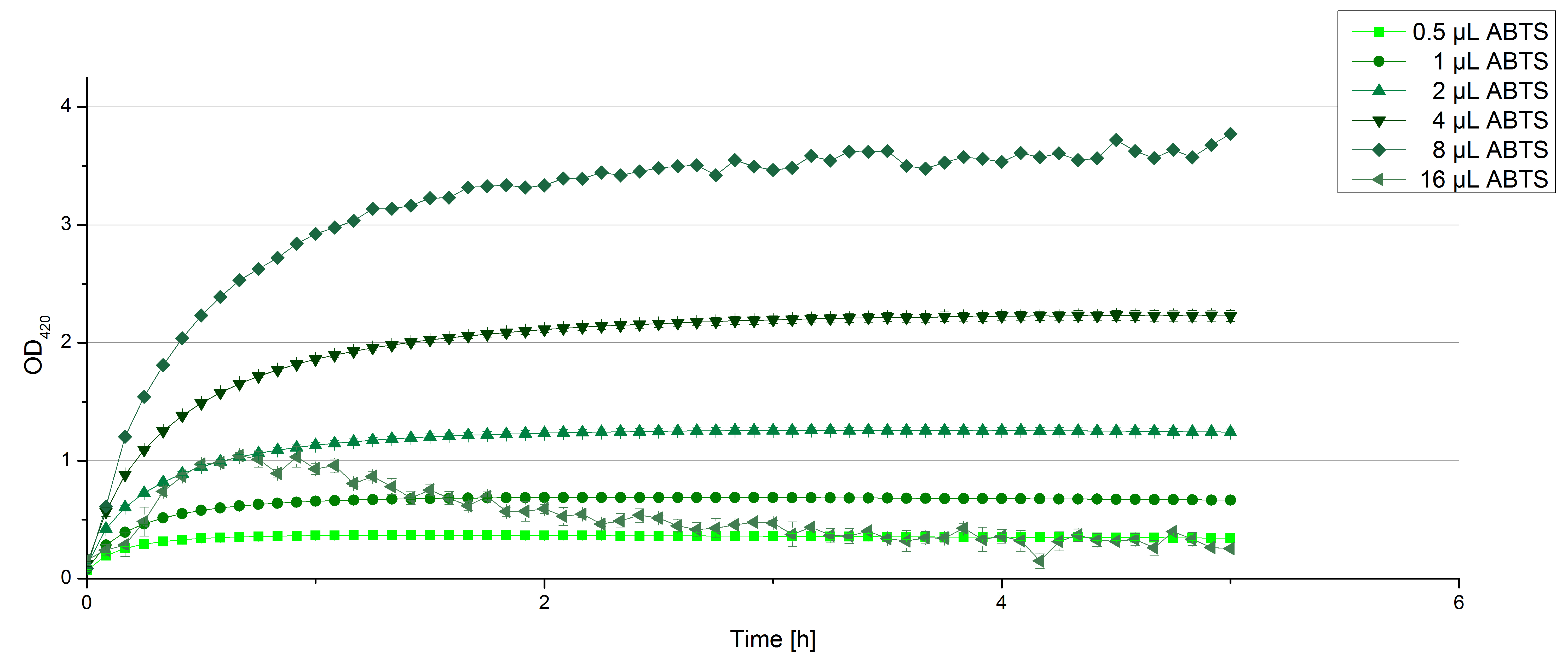
[[File:Bielefeld2012 activityKevin1508.jpg|thumb|left|Output of enzyme activity measurements via oxidized ABTS and OD<sub>420</sub>. Measured is the activity of [http://partsregistry.org/Part:BBa_K863005 ECOL], [http://partsregistry.org/wiki/index.php/Part:BBa_K863015 XCCL], [http://partsregistry.org/wiki/index.php/Part:BBa_K863010 TTHL], [http://partsregistry.org/wiki/index.php/Part:BBa_K863020 BHAL] and [http://partsregistry.org/wiki/index.php/Part:BBa_K863000 BPUL]. ''E. coli KRX'' (no plasmid) was used as negativ control and cells containing a plasmid with ligase was used for the control of induction. The binding buffer was as well tested as a negative control. ]] | [[File:Bielefeld2012 activityKevin1508.jpg|thumb|left|Output of enzyme activity measurements via oxidized ABTS and OD<sub>420</sub>. Measured is the activity of [http://partsregistry.org/Part:BBa_K863005 ECOL], [http://partsregistry.org/wiki/index.php/Part:BBa_K863015 XCCL], [http://partsregistry.org/wiki/index.php/Part:BBa_K863010 TTHL], [http://partsregistry.org/wiki/index.php/Part:BBa_K863020 BHAL] and [http://partsregistry.org/wiki/index.php/Part:BBa_K863000 BPUL]. ''E. coli KRX'' (no plasmid) was used as negativ control and cells containing a plasmid with ligase was used for the control of induction. The binding buffer was as well tested as a negative control. ]] | ||
* '''Team Substrate Analytic:''' | * '''Team Substrate Analytic:''' | ||
| - | ** Dominik | + | ** Dominik tried to regenerate the column. After that we measured new dilution series from all three estrogens. Once again all substrates left the column together with the solvent. That means the column is defect. |
| - | ** We ordered new C18-columns from Knauer and waited for | + | ** We ordered new C18-columns from Knauer and waited for their arriving. A Eurosphere and a Bluespehere column. |
===Thursday August 16th=== | ===Thursday August 16th=== | ||
Revision as of 00:03, 27 September 2012
Prologue
Starting the team
Beginning in january and february members of the former iGEM team from Bielefeld started seminars to inform interested students about synthetic biology, iGEM and the past Bielefeld projects. In March the final 2012 iGEM Bielefeld team was formed of 15 students and weekly meetings began. Our team was established and it was time to find a suitable project.
Find a project
The first weekly meeting were more like big group brainstorming and we discussed idea, which in some cases were totally different from each other. Everyone had to inform about ideas of others so that, in the end, we all could discuss together.
First project ideas were:
- the detection of multiresistent pathogens
- communication between bacteria and fungi using quorum sensing
- a bacterial hand warmer
- a possibility to detect and destroy mold fungus
- something about spontaneous combustion of hay bale
- an enzyme dispenser
For most of the ideas little information was available. For example spontaneous combustion of hay bales is probably a combination of the metabolisms of different microorganisms and fungus. After some reports in media and press about the environmental effects of steroid hormones, we decided to go for hormones. From the beginning our aim was not to detect but to degrade hormones. We found several possible ways for degradation as there are the hydrolysis of estradiol-derivates with sufatases and glucoronidases. But we thought the best way to degrade steroid hormones would be with the use of laccases. Laccases have the ability to radicalize aromatic rings and can therefore be used to degrade or polymerize a broad range of substances, such as steroid hormones, special insecticides, polycyclic aromatic carbohydrates and aromatic acids. In nature laccases are often used for degradation or polymerisation of lignin or pigments.
Molding together to a team
After we found our project idea we decided to have a get-to-know-weekend with some presentations about iGEM, important methods and ideas for human practices. We also held presentations about other possible iGEM projects to extend our horizon, as there were: e.g. RNA aptamers and magnetotactic bacteria. But the most important part of this weekend was the growing as a team. We realized that we all had one summer to work together, have fun together and most important to stand up together as a team.
Find the right one for the right job
Now it was time to organize the work and find a suitable task for everyone. In a developing team a lot of different jobs have to be done, e.g.:
- finding sponsors
- communication with the public
- human practices
- wiki- and homepage-design
- modelling
- a forum for exchange of information
- a joker, who entertains the team and lifts the mood
And finally lab work began, feel free to follow us in our weekly labjournal and have a look how our labwork, results and of course problems and their solutions, evolved.
Summary of Week 1
We began our time in the lab with the cultivation of Xanthomonas campestris B100 and E. coli BL21(DE3) to isolate the genomic DNA to do PCRs and purify the desired laccase ORFs. In Order to do this we at first had to designed the PCR-primers. We decided, that the forward primers had to included the prefix, the T7 promotor, the RBS and the first 20 bases of the gene of interest and the reverse primers should consist of the last 20 bases of the gene of interest, a His-tag, followed by two stop codons and the suffix. Furthermore we started making preparations for the Student Academy. The Students Academy is a week-lasting summer school (9th to 13th of July) we got the chance to take part in organizing it. It is distinguished with a lot of presentations and lessons for pupils, but also guided experiments they had to do by themselves. Therefore the general idea for our experimental part was to give the students an understanding of the principle methods in biotechnology and synthetic biology by using fluorescent proteins. So the first step was to searched the Parts Registry for two plasmids with different fluorescent proteins and antibiotic resistances, respectively.
Week 1 (04/30 - 05/06/12)
Contents |
- Start of our WET LAB time.
Weekly Seminar
- Do we want to order strains of Trametes versicolor and Trametes villosa?
- Gathering information about signal sequences in yeast
- Decision to create a database, so that we can easily number and inscribe our lab results
- Decision to arrange a summer school for pupils in their last year before the final exams
- Discussion about how to meet a member of the german [http://www.bundestag.de/ Bundestag] (the german parliament)
Monday April 30th
- Team Student Academy:
- We got the chance to organize one part of the first school academy “synthetic biology/ biotechnology” at the CeBiTec of University Bielefeld by arranging experiments for the pupils and by presenting us and the iGEM competition. For the experimental part our general idea was to give them an understanding of principle methods in biotechnology / synthetic biology by using fluorescent proteins. We planned the following experiments:
- Plasmid isolation of RFP/GFP from a liquid culture.
- Transformation of a plasmid mixture consisting of two different fluorescent proteins (e.g. RFP and GFP) and different antibiotic resistances into E. coli KRX. It will be plated out on LB agar plates without antibiotics and on plates containing one of the two antibiotics, which are present on the plasmids. This way we can demonstrate the effect of antibiotics as selective pressure.
- We got the chance to organize one part of the first school academy “synthetic biology/ biotechnology” at the CeBiTec of University Bielefeld by arranging experiments for the pupils and by presenting us and the iGEM competition. For the experimental part our general idea was to give them an understanding of principle methods in biotechnology / synthetic biology by using fluorescent proteins. We planned the following experiments:
- Team Cloning of Bacterial Laccases:
- Before our lab time started we sent requests for different plasmids with the desired laccase genes to working groups, which have already worked with laccases we are interested in. Sadly just one working group responded to us. We got answer for a vector with the laccase-ORF [http://www.ncbi.nlm.nih.gov/protein/194015788 CotA] from Bacillus pumilus ATCC7061 and an ampicillin resistance from the Swiss Federal Laboratories for Materials Science and Technology, Laboratory for Biomaterials in Switzerland. They promised to send us the plasmid pBpL6. [http://www.biomedcentral.com/1472-6750/11/9 More information...] For an uniformly labeling we will further call this laccase BPUL.
- In a [http://www.ncbi.nlm.nih.gov/pubmed/21790191 publication] we found a research group who worked with the laccase [http://www.ncbi.nlm.nih.gov/protein/21230052 CopA] from Xanthomonas pv. campestris ATCC33913. Luckily the sequence of this laccase is the same in Xanthomonas campestris pv. campestris B100 which we got from a research group at our university. We will call this laccase XCCL in our wiki from now on.
- We found a [http://www.springerlink.com/content/x33205rp257397kr/ publication] which described the laccase [http://www.ncbi.nlm.nih.gov/protein/85674340 CueO] from E. coli W3110. After blasting this laccase we found out that E. coli BL21(DE3) has this laccase, too. We decided to isolate the laccase from E. coli BL21(DE3) because this strain is available in our lab.
- We generated new competent E.coli KRX cells.
- For the extraction of genomic DNA we cultivated Xanthomonas campestris B100 and E. coli BL21(DE3). The bacterial strains we got from a working group at our University. After cultivation we isolated the genomic DNA. The DNA was needed as template for PCRs to purify the wanted laccase ORFs.
- Primer design for the isolation of laccases from genomic DNA of Xanthomonas campestris B100 and E. coli BL21(DE3) and from plasmid pBpL6 Bacillus pumilus ATCC7061.
- In a team meeting earlier in the project we decided that we want to express the laccases under control of an inducible T7 promoter so we can choose the time point of induction. Additionally we decided that we want to use a c-terminal His-tag. Therefore the forward primers were designed with T7 promoter, RBS and the first 20 bases of the wanted gene. The reverse primers were designed with the last 20 bases of the wanted gene without the stop codon, a His-Tag and two stop codons. Primers: Xcc_LAC_FW_T7, Xcc_LAC_RV_HIS, E.coli_LAC_FW_T7, E.coli_LAC_RV_HIS, B.pumi_LAC_FW_T7 and B. pumi_LAC_RV_HIS
Tuesday May 1th
- Team Student Academy:
- Searching for two plasmids with different fluorescent proteins and antibiotic resistance in parts registry. Found [http://partsregistry.org/Part:BBa_J04450 BBa_J04450], a Plasmid with RFP and chloramphenicol resistance (but lacI and CAP sensitive), [http://partsregistry.org/Part:BBa_J23100 BBa_J23100], a plasmid with RFP and ampicillin resistance and [http://partsregistry.org/wiki/index.php?title=Part:BBa_I13522 BBa_I13522], a Plasmid with GFP and ampicillin resistance in Kit Plate 2011.
- Team Database:
- We decided to create a database, so that we can easily number and inscribe our lab results and that everybody has the chance to find out witch results the other groups has so far.
- Starting to look for possibilities to design such a database.
Wednesday May 2th
- Team Activity Test: Good morning everybody and welcome to the labjournal of Team Activity Tests. Today we started our work with some literature research about enzyme activity tests, laccases and its substrates. So today was filled with online research, reading papers and collecting information about the laccases our team decided to use.
Thursday May 3th
- Team Cloning of Bacterial Laccases:
- After the vector with the laccase gene bpul from Bacillus pumilus arrived, we transformed it into the competent E. coli KRX to have a larger amount of vector. The protocol we used was as followed:
- The electroporation setup: U = 2,5 kV, C = 25 µF and R = 400 Ω
- Since we did not know the efficient of our competent KRX we used two different E.coli volumes for the transformation, 50 µL and 100 µL. We gave 50 µL 10% glycerol to the reaction tubes with 1 µL of the vector DNA (Bacillus pumilus). After the transformation we plated them into ampicillin plates.
- PCR with the Xanthomonas campestris B100 and E. coli BL21(DE3) genomic DNA to isolate the laccases. Therefore we used the primers Xcc_LAC_FW_T7, Xcc_LAC_RV_HIS, E.coli_LAC_FW_T7 and E.coli_LAC_RV_HIS which are listed under Materials.
- After the vector with the laccase gene bpul from Bacillus pumilus arrived, we transformed it into the competent E. coli KRX to have a larger amount of vector. The protocol we used was as followed:
Friday May 4th
Team Cloning of Bacterial Laccases: We did Colony PCR on the transformed the Bacillus pumilus CotA plasmid. Unfortunately the control with colony PCR didn't work. So we just picked some colonies for plasmid isolation in the hope that on the AMP plate were no false positives colonies.
Summary of Week 2
hier eine Zusammenfassung
Week 2 (05/07 - 05/13/12)
Contents |
Weekly Seminar
- Found our first sponsors: [http://corporate.evonik.com/en/Pages/default.aspx Evonik], [http://www.biocircle.com/en-ca/ BioCircle] and [http://www.merckgroup.com/en/index.html Merck], now treaties have to be created and signed.
- Julia V. is working on the database.
- Decision to organize a waffle sale to fill up our petty cash.
- Gabi and Isabel are designing a poster for the waffle sale.
- For our human practices we wanted to find a sociology student, willing to think about bioethics, but did not succeed yet.
- Our video is nearly done, it is cut and only needs be underlain with music.
Monday May 7th
- Team Student Academy:
- First transformation of [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_I13522 BBa_I13522] and plating on selective agar. Result: We got little colonies. There weren’t any green colonies and only some pale red fluorescent colonies.
- Team Cloning of Bacterial Laccases:
- More PCRs of laccase genes xccl from Xanthomonas campestris pv. campestris B100 and ecol from E. coli BL21(DE3) with the isolated genomic DNA as template and Xcc_LAC_FW_T7 / Xcc_LAC_RV_HIS and E.coli_LAC_FW_T7 / E.coli_LAC_RV_HIS primer pairs.
- Since we wanted to screen and characterize laccases from different bacteria we had to order the bacterial strains which weren't available at Bielefeld University from [http://www.dsmz.de/|DSMZ]. Below is a list of the ordered strains and the laccases we want to isolate from these strains.
- [http://www.ncbi.nlm.nih.gov/protein/46197298 Laccase] from [http://www.dsmz.de/catalogues/details/culture/DSM-7039.html?tx_dsmzresources_pi5%5BreturnPid%5D=304|Thermus thermophilus HB27] (look [http://www.ncbi.nlm.nih.gov/pubmed/15999224 here] for a publication to this laccase)
- [http://www.ncbi.nlm.nih.gov/protein/10174701 BH2082] from [http://www.dsmz.de/catalogues/details/culture/DSM-18197.html?tx_dsmzresources_pi5%5BreturnPid%5D=304|Bacillus halodurans C-125] (look [http://www.ncbi.nlm.nih.gov/pubmed/15293032 here] for a publication to this laccase)
- We ordered [http://www.dsmz.de/catalogues/details/culture/DSM-40069.html?tx_dsmzresources_pi5%5BreturnPid%5D=304|S. lavendulae sp. lavendulae ATCC 14158]. Originally we wanted the strain Streptomyces lavendulae REN-7 but this strain isn't available at DSMZ. So we now hope that the laccase gene [http://www.ncbi.nlm.nih.gov/nuccore/23491745 STSL] from Streptomyces lavendulae REN-7 is similar to that from S. lavendulae sp. lavendulae ATCC 14158 because there's no DNA sequence for the laccase from this strain available. [http://www.ncbi.nlm.nih.gov/pubmed/14586105 publication]
- We wanted the laccase [http://www.ncbi.nlm.nih.gov/protein/182434812 EpoA] from Streptomyces griseus IFO 13350 (for the publication look [http://jb.oxfordjournals.org/content/133/5/671.full.pdf here]. This strain was not available so we ordered [http://www.dsmz.de/catalogues/details/culture/DSM-40236.html?tx_dsmzresources_pi5%5BreturnPid%5D=304| Streptomyces griseus ATCC 10137]. Unfortunately for this strain are no blast results after blasting the [http://www.ncbi.nlm.nih.gov/protein/182434812 laccase] from Streptomyces griseus IFO 13350 against database. So we decided to make primers for the laccase sequence from Streptomyces griseus IFO 13350 in the hope that the sequences are similar enough to get a PCR product.
- Team Modeling:
- Looking for suitable software and enzymkinetics to model the degradation of our substrates with the different laccases. Finding the Michaelis-Menten kinetics and matlab.
Tuesday May 8th
- Team Student Academy:
- Repetition of the transformation didn’t change the result. We made a liquid culture of [http://partsregistry.org/Part:BBa_J04450 BBa_J04450], but it did not fluoresce. Searching for mistakes and alternatives. Maybe competent cells are not that good and in case of RFP the lacI sensitivity could be the problem.
- Team Cloning of Bacterial Laccases:
- After some empty agarose gels we finally isolated the laccase gene bpul from Bacillus pumilus ATCC7061 as PCR product with the desired overhanging ends. As template we used the plasmid we got from the Swiss working group.
Wednesday May 9th
- Team Activity Test: From the information we collected during our literature research we created a protocol for our first experiments. We decided to check the activity via a photometer. The one we may use here at the Cebitec is a Tecan Microplate reader. Check protocols for further information. If oxidized by laccase, ABTS can me measured at 420 nm. Also we found out that sodium acetate buffer (100 mM / pH 5) would give an optimal environment to our enzyme. So let´s have a look at our protocol:
- Initial laccase activity test:
- 100 mM sodium acetate buffer, pH 5.0
- 5 mM ABTS
- 8 U laccase
- ad 200 µL deionized H20
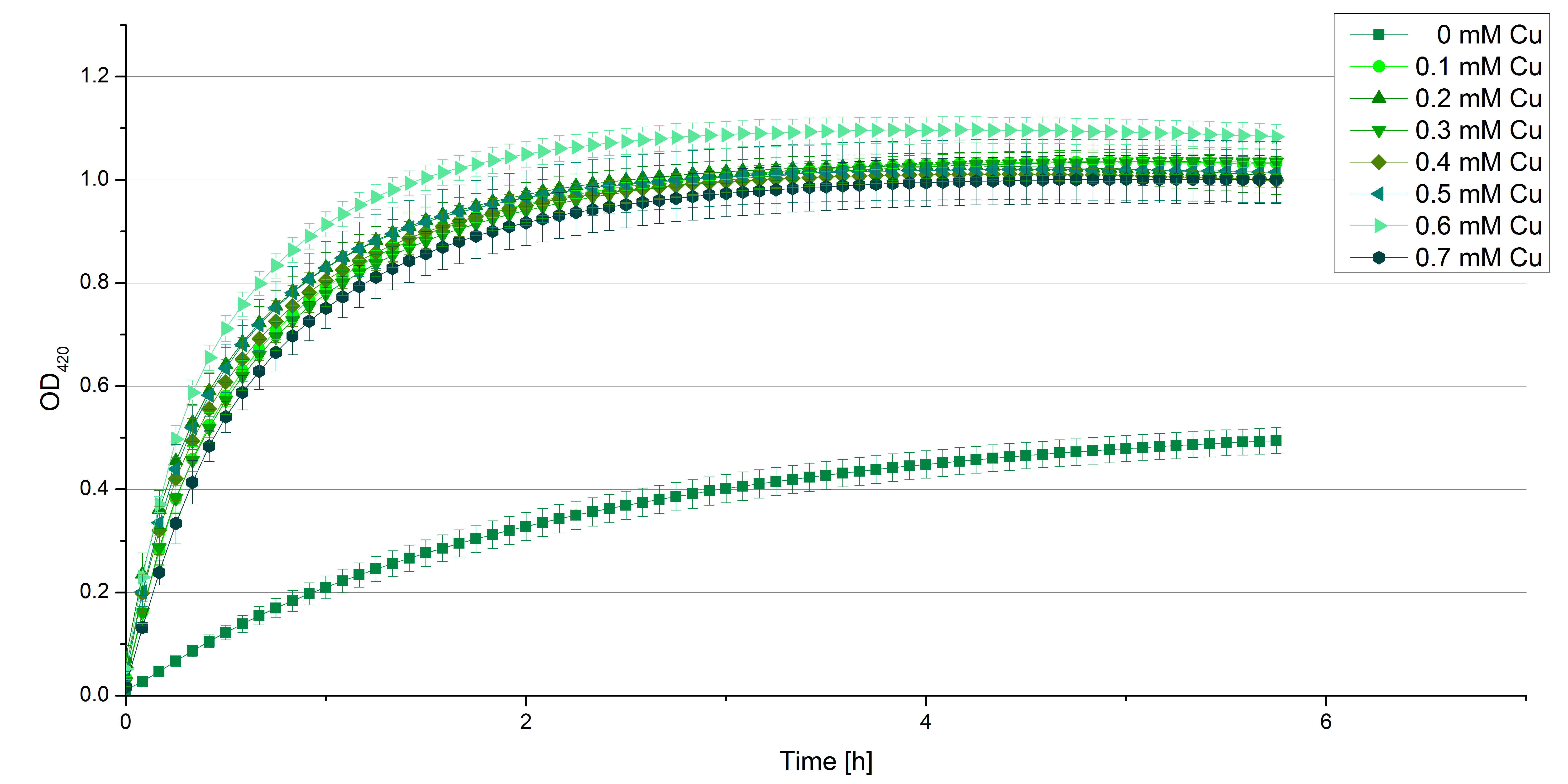
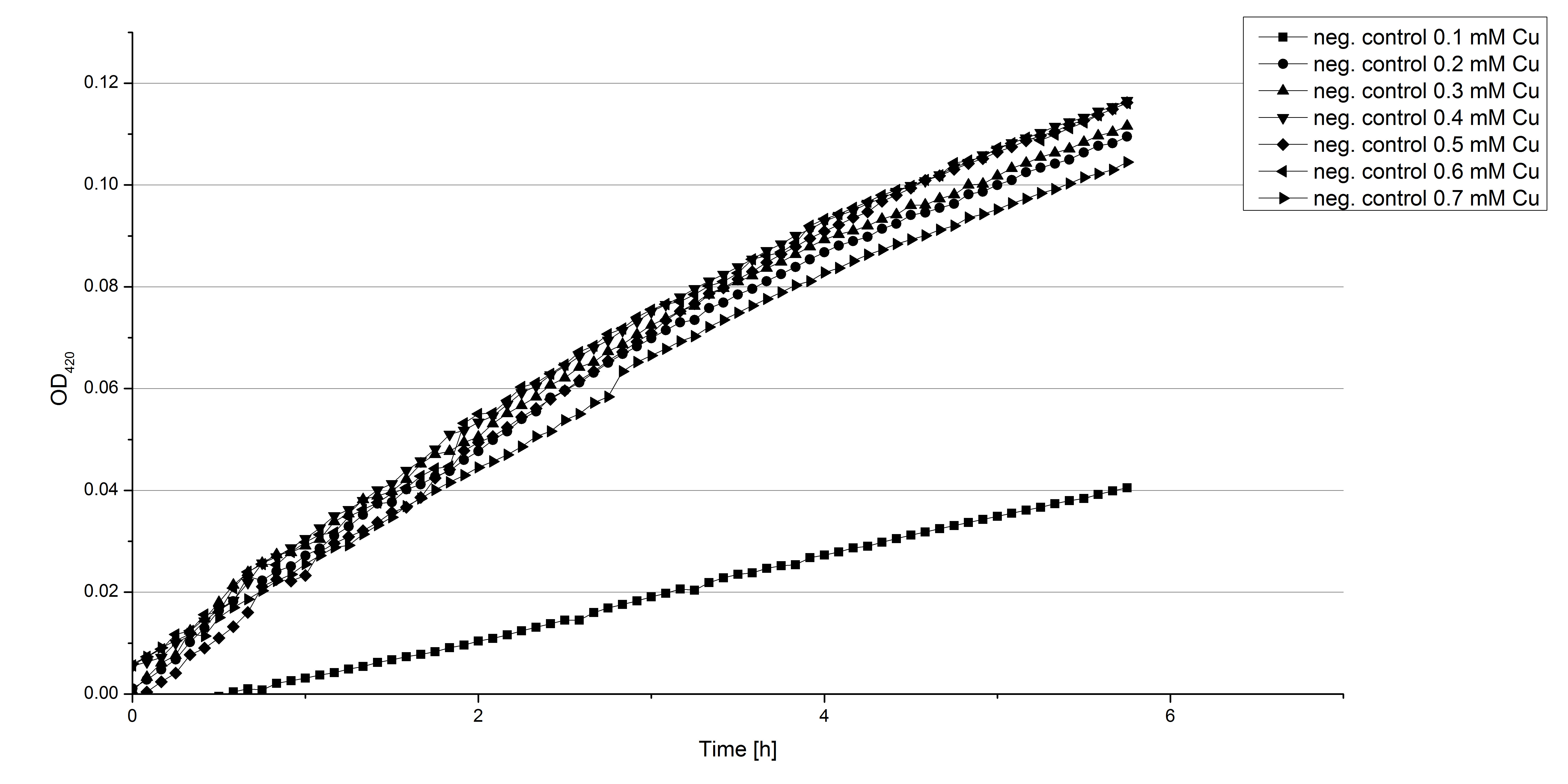
- Also we talked about further characterization after accomplishing the first experiments and confirming that the used concentrations are a good choice. We are planning to buy and characterize the laccase from T.versicolor (TVEL0), to have a comparison to our future recombinant laccases. That laccase we are going to analyze in sodium acetate buffers that are adjusted to pH 1, 3, 5, 7 and 9. Further we are going to analyze the effect of different temperatures on the enzymes activity. For that we will first do some more research on the temperatures of the waste water in clarification plants here in Germany. Also we found out that an addition of copper does enhance the laccases activity, so we are going to do some measurements with copper concentrations from 0.1 mM to 0.5 mM in each sample. This seems like some great experiments for the start, so next we are going to order what we need to do the measurements.
- Initial laccase activity test:
- Team Database
- Finding a first initial design
Thursday May 10th
- Team Student Academy
- Testing the competent cells by transformation of pUC19. The transformation did not work that good, so that we produced new ones.
- Team Cloning of Bacterial Laccases
- We got the ordered strains from [http://www.dsmz.de/ DSMZ]. So we did PCR on Thermus thermophilus genomic DNA. First we dissolved some of the lyophilized powder in water and for opening the cells we boiled them for a few minutes. The primers we used were T.thermo_LAC_FW_T7 and T.thermo_LAC_RV_HIS to get the laccase with the same overhangs described in Monday April 30th. Finally with additional DMSO and GC-buffer we had a product of the GC-rich laccase.
Friday May 11th
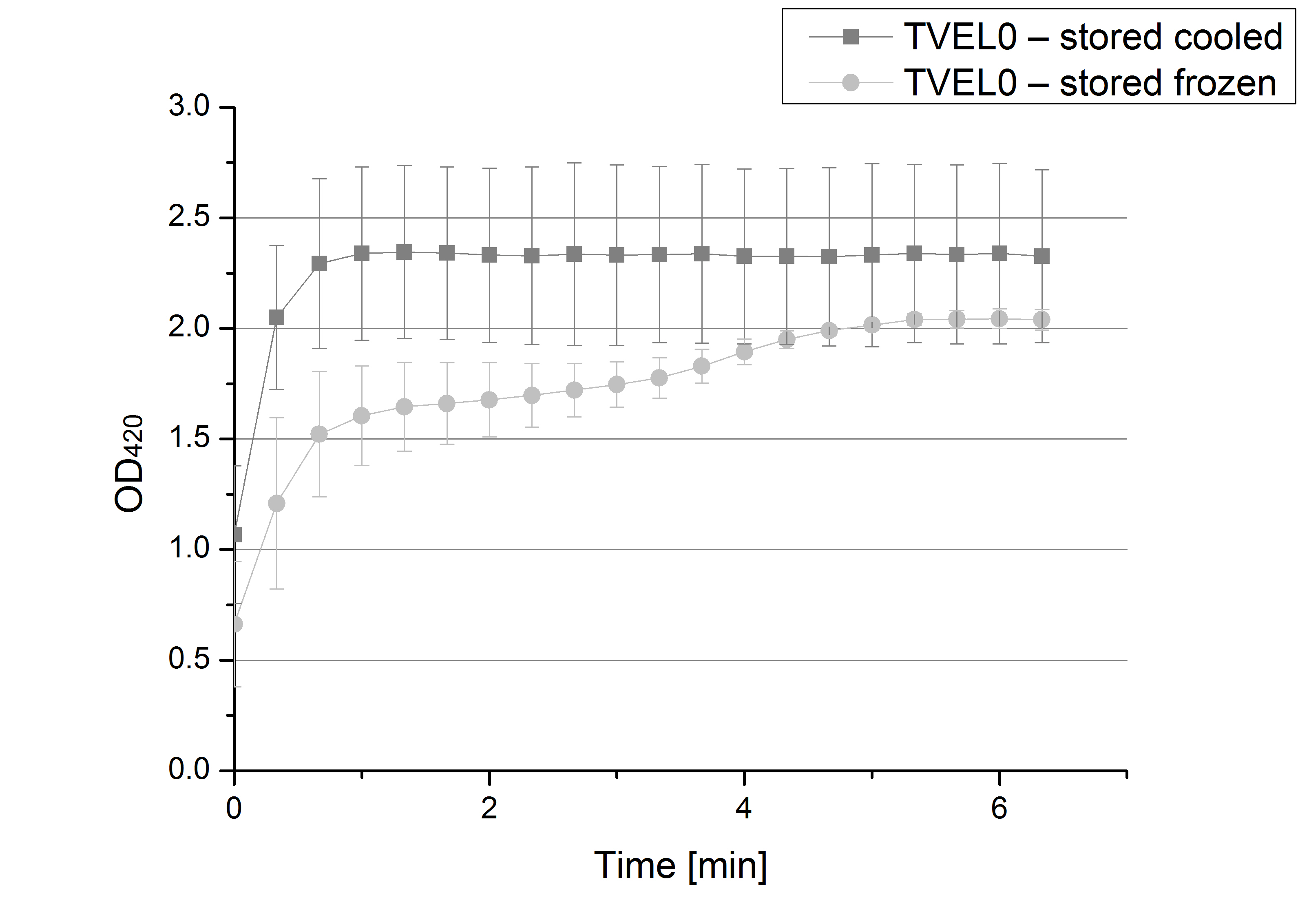
- Team Activity Tests: For some pre test and characterization for our future laccase activity standard we ordered [http://www.sigmaaldrich.com/catalog/product/sigma/53739?lang=de®ion=DE laccase] from Trametes versicolor. As well we had to order a substrate that the laccase could use to demonstrate its abilities. According to the literature [http://www.sigmaaldrich.com/catalog/product/sigma/a1888?lang=de®ion=DE ABTS] is a well working substrate to characterize oxidizing enzym activity. So we ordered.
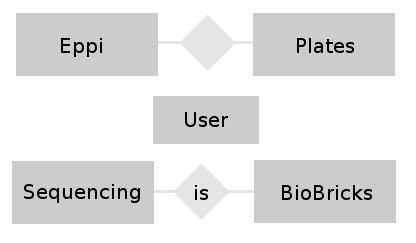
- Team Database: Conceptual Design: In this phase of the design we try to find relations between the different tables and build a entity-relationship-model. This model helps to visualize the entities (our different tables) together with their attributes (the entries belonging to a table) and the relationship between these entities. The next figure shows a very easy E-R-Model.
Summary of Week 3
hier eine Zusammenfassung
Week 3 (05/14 - 05/20/12)
Contents |
Weekly Seminar
- first lab service: Robert
- our GFP, which we wanted to use for the summer school for pupils, does not work.
- first competent cells have to be made: Julia S. and Robert.
- Decision to buy a commercial laccase to establish the analytics and the enzyme activity tests.
- our scientific exposé has to be translated into english: Malak
- Last planning for our waver sell.
- Julia S. is creating a vector for Pichia pastoris and is now searching for usable sequences.
- The [http://www.bmbf.de/en/index.php BMBF] invites all german iGEM teams to Berlin to attend at the Biotechnologie2020+ strategy process.
Monday May 14th
Tuesday May 15th
- Team Database:
- Creating the logically design for the database. For example for our table 'Eppi' we opt for the following logic:
| eppi ( | ID INT(11), |
| DataID INT(11), | |
| Caption TEXT, | |
| Content TEXT, | |
| Experiment VARCHAR(255), | |
| Where2 VARCHAR(255) , | |
| Location TEXT, | |
| Coments TEXT, | |
| Empty ENUM ('N', 'J'), | |
| Create_User INT(11), | |
| Creat_Date DATETIME, | |
| Edit_Date DATETIME, | |
| Delete ENUM ('N', 'J') | |
| Delete_User INT(11), | |
| Delete_Date DATETIME ) |
Wednesday May 16th
- Team Cloning of Bacterial Laccases:
- For cloning our laccases we need pSB1C3 backbone. Therefore we we transformed [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] (pSB1C3 with RFP) in competent KRX cells.
Thursday May 17th
- Team Cloning of Bacterial Laccases: Plasmid isolation of <partinfo>BBa_J04450</partinfo>.
- Team Modeling:
- Meeting Mrs. Lutter, a mathematics prof. of our course of studies and looking for our first model of a metabolic pathway, finding out, that we don't need such a complex model. Start thinking that we want and what we need.
Friday May 18th
- Team Activity Tests: Our [http://www.sigmaaldrich.com/catalog/product/sigma/53739?lang=de®ion=DE T.versicolor laccase] and the [http://www.sigmaaldrich.com/catalog/product/sigma/53739?lang=de®ion=DE ABTS] arrived! We couldn´t wait to start, so we set up the stock solutions we will need, such as sodium acetat buffer (pH 5), 10 mM ABTS and deluted laccase.
Saturday May 19th
Sunday May 20th
Summary of Week 4
hier eine Zusammenfassung
Week 4 (05/21 - 05/27/12)
Contents |
Weekly Seminar
- Lab service: Isabel
- We try to establish a collaboration with the iGEM team from SDU-Denmark.
- Got our distribution kits
- First successful cloning and cultivations.
- Who wants to be a summer school teacher?
- We will not travel to the ACHEMA because only local teams are invited.
- Do we want to participate in the Biolympics? (It's a sports party with fun organized by the [http://bts-ev.de/ bts]).
Monday May 21st
- Team Cloning of Bacterial Laccases:
- We wanted to clone our laccase PCR products xccl and ecol in pSB1C3 backbone. Therefore we did some restriction digests on the PCR products and the vector [http://partsregistry.org/Part:BBa_J04450 BBa_J04450].
- Team Modeling: Our aims for modeling:
- model the expression of the laccases in the organisms.
- model the activity of our enzymes.
- model the interesting parts of a clarification plant (the part witch are interesting for our cleaner.
Tuesday May 22nd
- Team Cloning of Bacterial Laccases:
- Ligation of the digested PCR products in pSB1C3 backbone and transformation in KRX electro-competent cells.
Wednesday May 23rd
- Team Student Academy:
- Repetition of the transformation of [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_I13522 BBa_I13522] with new competent E. coli KRX cells. Got intense red fluorescing colonies but no green fluorescing colonies. Made a backup of E. coli KRX [http://partsregistry.org/Part:BBa_J04450 BBa_J04450].
- Asking for other plasmids containing GFP at the working groups of our University.
- Team Cloning of Bacterial Laccases:
- After there were no colonies on our pSB1C3 + xccl(T7)_His (Xanthomonas campestris) transformation plate we did the transformation with the same ligation preparation again. The other ligation with pSB1C3 + ecol(T7)_His (E. coli) showed colonies so we started colony PCRs to find positive colonies. Sadly the colony PCRs showed no products but the problem was that we just had the long overhang primers (E.coli_LAC_FW_T7 and E.coli_LAC_RV_HIS ). Therefore we ordered the F and R primers.
Thursday May 24th
- Team Student Academy:
- Made a liquid culture of E. coli KRX with [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] at 30 °C. There was no fluorescence.
- Transformation of [http://partsregistry.org/Part:BBa_J23100 BBa_J23100] into E. coli KRX. Also got intense red fluorescing colonies.
- Team Activity Tests:
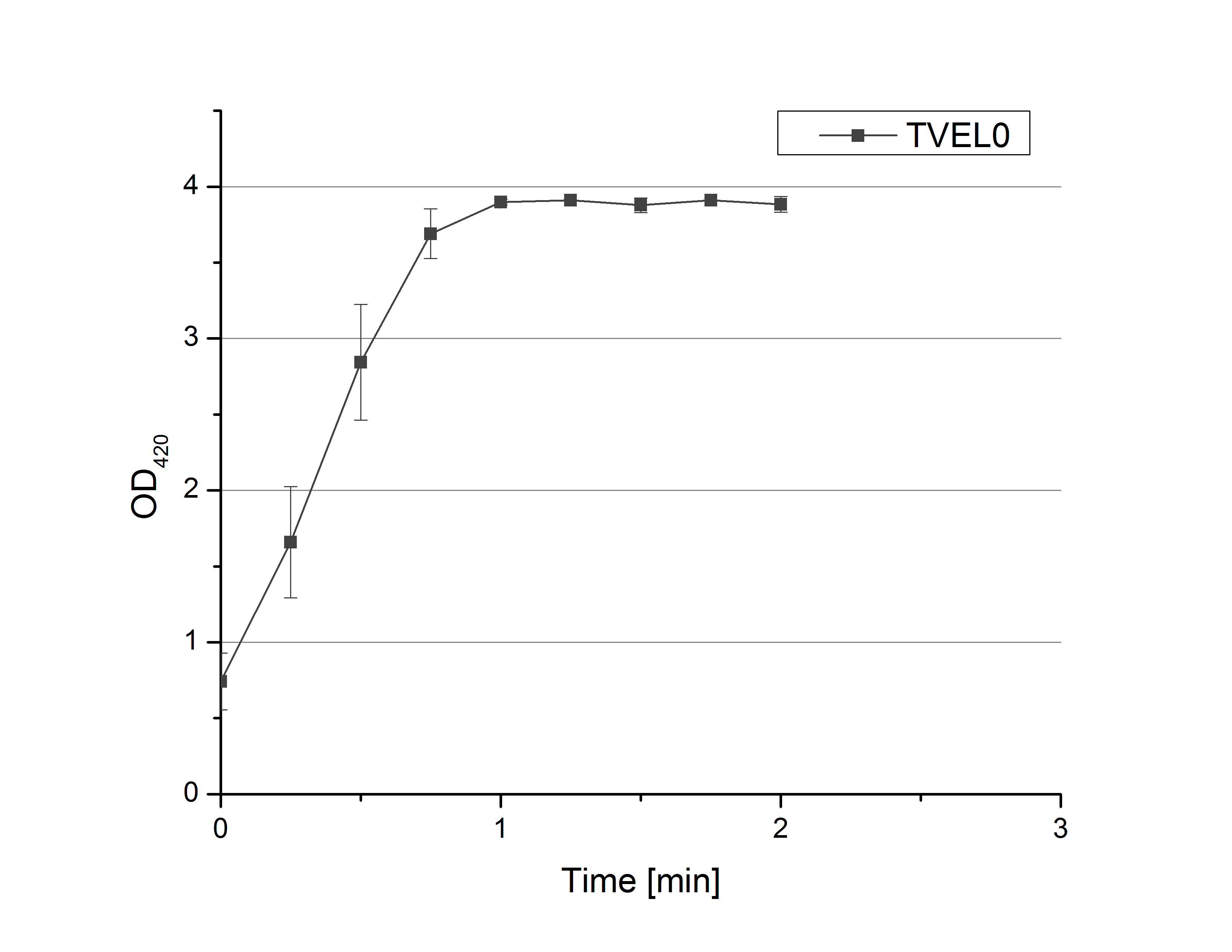
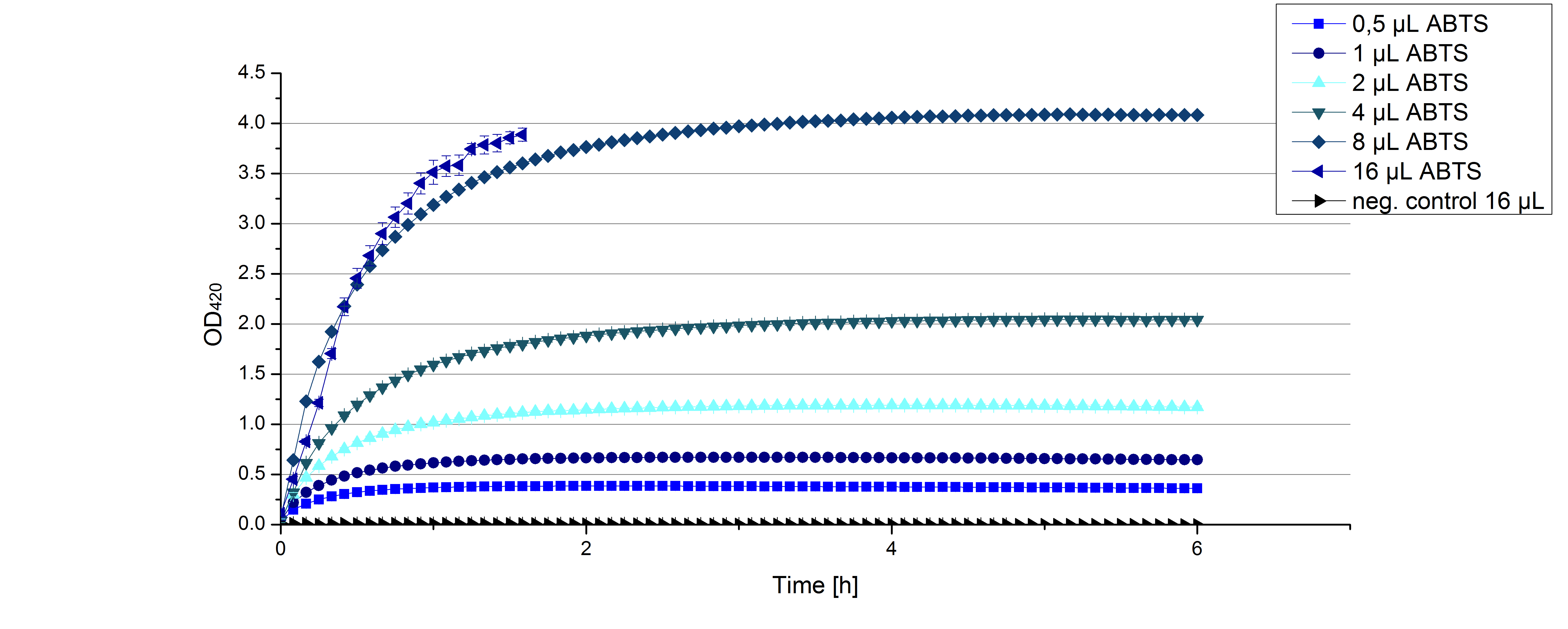
- On our schedule today was testing our bought laccases (from now on called [http://www.sigmaaldrich.com/catalog/product/sigma/53739?lang=de®ion=DE TVEL0]) and improving our protocol in case it won't work. We got familiar with the [http://www.greinerbioone.com/UserFiles/File/PDFCatalogue/e_2_6_96_Well_Microplates.pdf flat bottom microplates] and got a briefing into using the Tecan correctly. We started with the following setup: 100 mM sodium acetate (pH 5), 5 mM ABTS, 8 U TVEL0 laccase, ad 200 µL with deionized water. Then we measured the absorption at 420 nm every 15 seconds over a time period of 2 minutes. Immediately our samples turned dark blue but unfortunately the changeover was out of range to be detected by Tecan. With this information we needed to reduce the laccase concentration to get measurable results. We have chosen to try another measurement with 0.1 U TVEL0 laccase and it worked! The result was a nice saturation curve but it reached a too high, because not good measurable, optimum within 1 minute (Fig. 1). To avoid the loss of important data in the beginning of the reaction and to reduce the saturation OD we decided to slow everything down by using less ABTS.
Friday May 25th
- Team Cloning of Bacterial Laccases:
- We had to do the PCR on T. thermo laccase again because after the purification of the last PCR product the DNA amount was very low.
- Team Student Academy:
- Made a liquid culture of E. coli KRX with [http://partsregistry.org/Part:BBa_J23100 BBa_J23100] at 30 °C. Result: There was no fluorescence.
- Team Database:
- Yeah we have a login!!
Saturday May 26th
Sunday May 27th
- Team Activity Test:
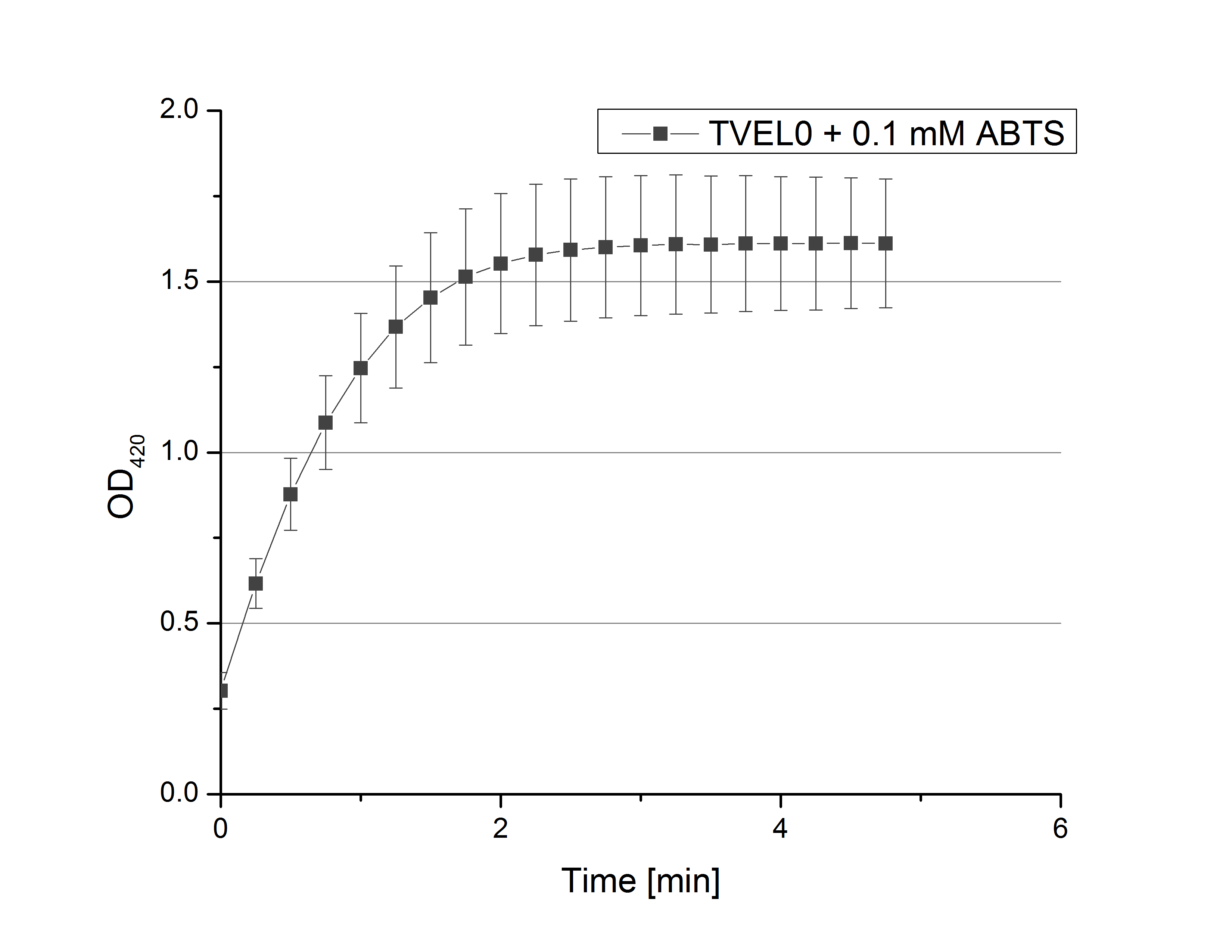
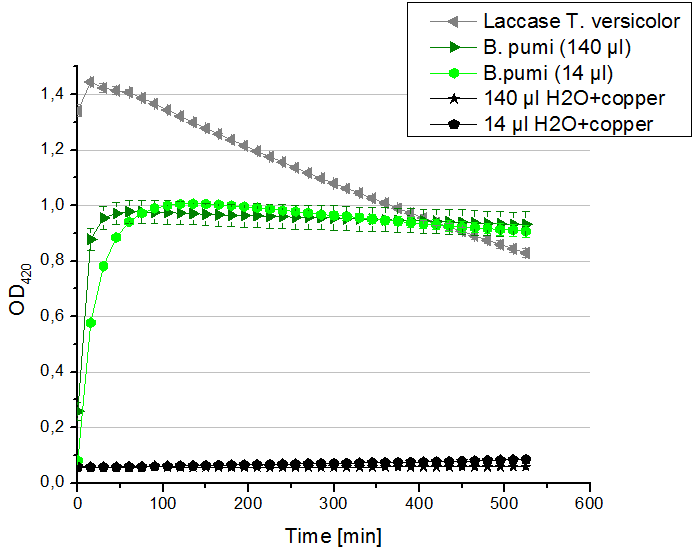
Remember our first activity measurements? The activity was awesome and reached quickly its saturation. So today we decided to have a lazy Sunday and reduce absorption maximum and reaction speed, too. For this we used 0.1 mM ABTS instead of 5 mM ABTS and we got good results. The saturation reached its maximum at a OD420 of ~1.6 after roundabout 2 minutes (see Fig. 2). Taking together we halved the maximal OD420 and doubled the reaction time until maximum is reached with using 0.1 mM ABTS instead of 5 mM ABTS.
Summary of Week 5
hier eine Zusammenfassung
Week 5 (05/28 - 06/03/12)
Contents |
Weekly Seminar
- We found our next sponsor: [http://www.analytik-jena.de/en/ Analytik Jena AG].
- all sub-teams have to present their results and problems next monday.
- Things to do in the lab:Site directed mutagenisis, cultivation, purification.
- Kevin will bring a white board for better communication in the lab.
- Julia presented the first screenshots of her database.
- Derya will be responsible for sequencing from now on.
- Collaborations: Nadine will contact SDU-Denmark and Darmstadt.
- [http://www.cas.uni-muenchen.de/veranstaltungen/tag_synth_bio_2012/index.html CAS conference for Synthetic Biology] (23th - 25th of july) in Munich, Miriam will read up on travel and acccommodation expenses
- [http://www.biotechnologie2020plus.de/ strategy process "Biotechnologie2020+"] in Berlin, Gabi, Timo and Robert will represent the team
- press release: last changes are in progress
Monday May 28th
- Team Student Academy:
- Made liquid cultures of E. coli KRX with [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] and with [http://partsregistry.org/Part:BBa_J23100 BBa_J23100]. Result: Intense red fluorescence. We made a glycerol stock and a plasmid isolation
- Team Cloning of Bacterial Laccases:
- Digest of ecol(T7)_HIS, xccl(T7)_His and bpul(T7)_His and [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] (pSB1C3 + RFP) with the same restriction enzymes, ligation of the digested products and transformation in E. coli KRX cells.
- Since we have less bpul(T7)_His and tthl(T7)_His DNA, we set an PCR from the remaining PCR approach.
Tuesday May 29th
- Team Student Academy:
- E. coli Mach1 with pMTE cp46 His received from the working group “Fermentation Engineering”, University Bielefeld. Plasmid contains genes for GFP and kanamycine resistance. We plated it and made a liquid culture at 37°C. Result: There was an intense fluorescence. We made a glycerol stock and a plasmid isolation.
- Team Cloning of Bacterial Laccases:
- We did colony PCRs on the transformations from the day before. We got product for every transformation approach so we plated the positive colonies on new plates to make plasmid isolation. So hopefully in some days we have the plasmids with the E. coli laccase, the Xanthomonas campestris laccase and the B. pumilus laccase with the inducible T7 promoter and a His-tag.
- Team Activity Tests:
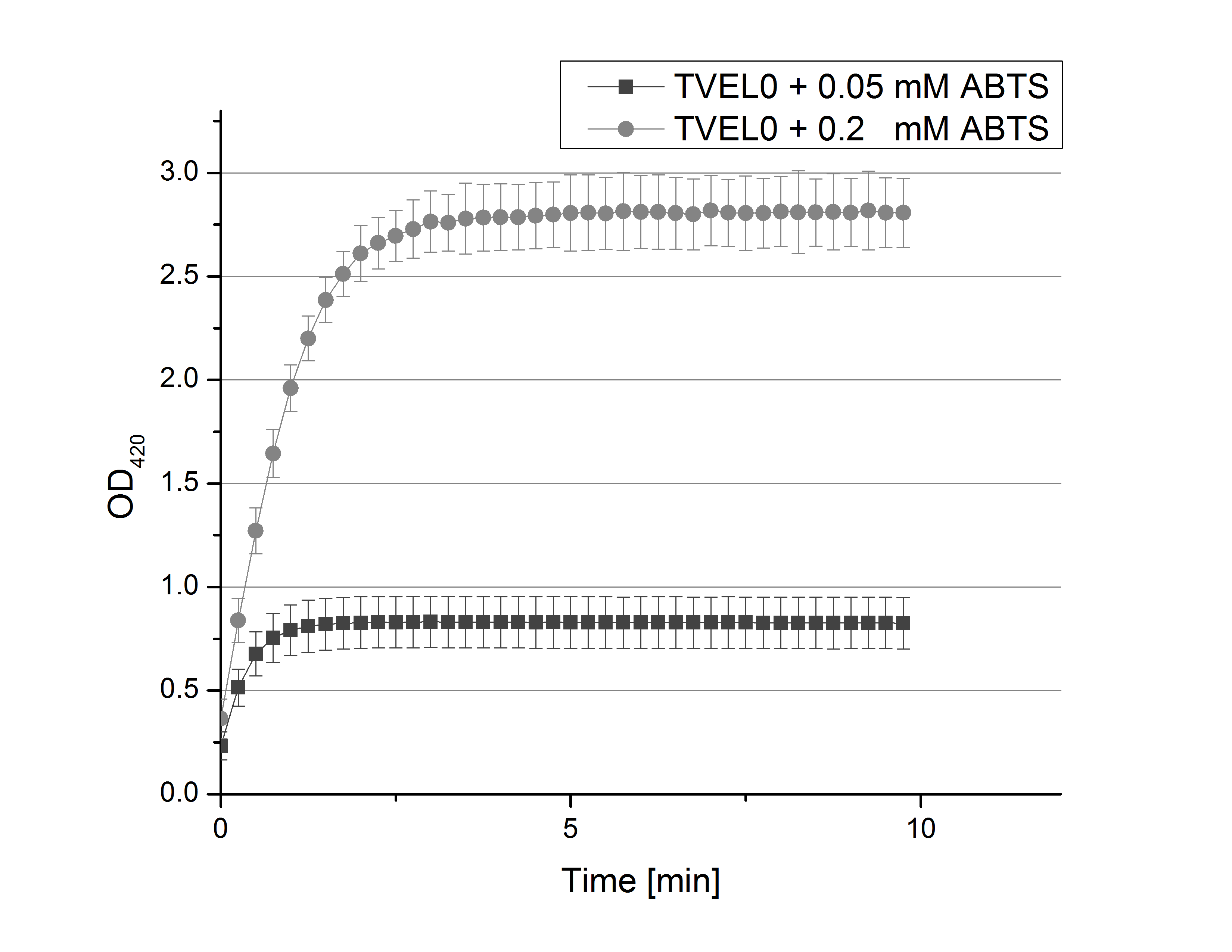
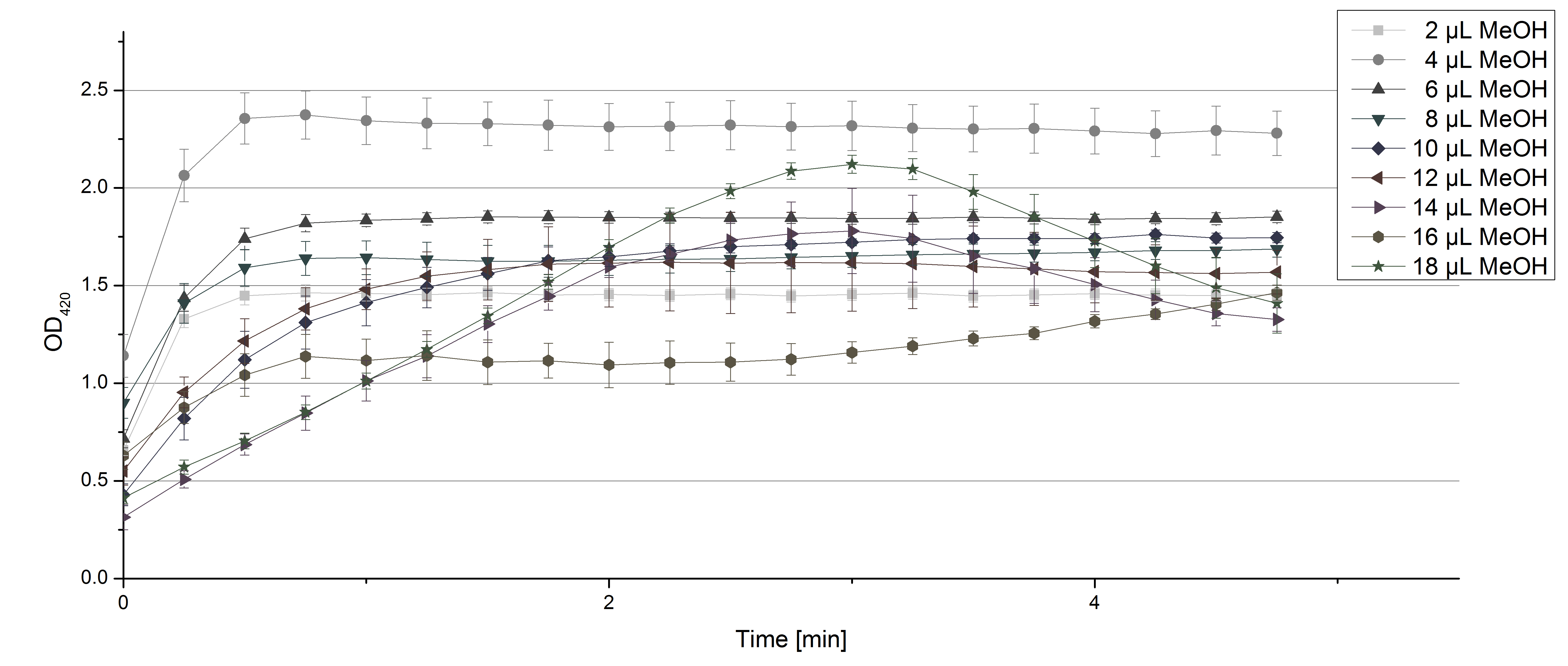
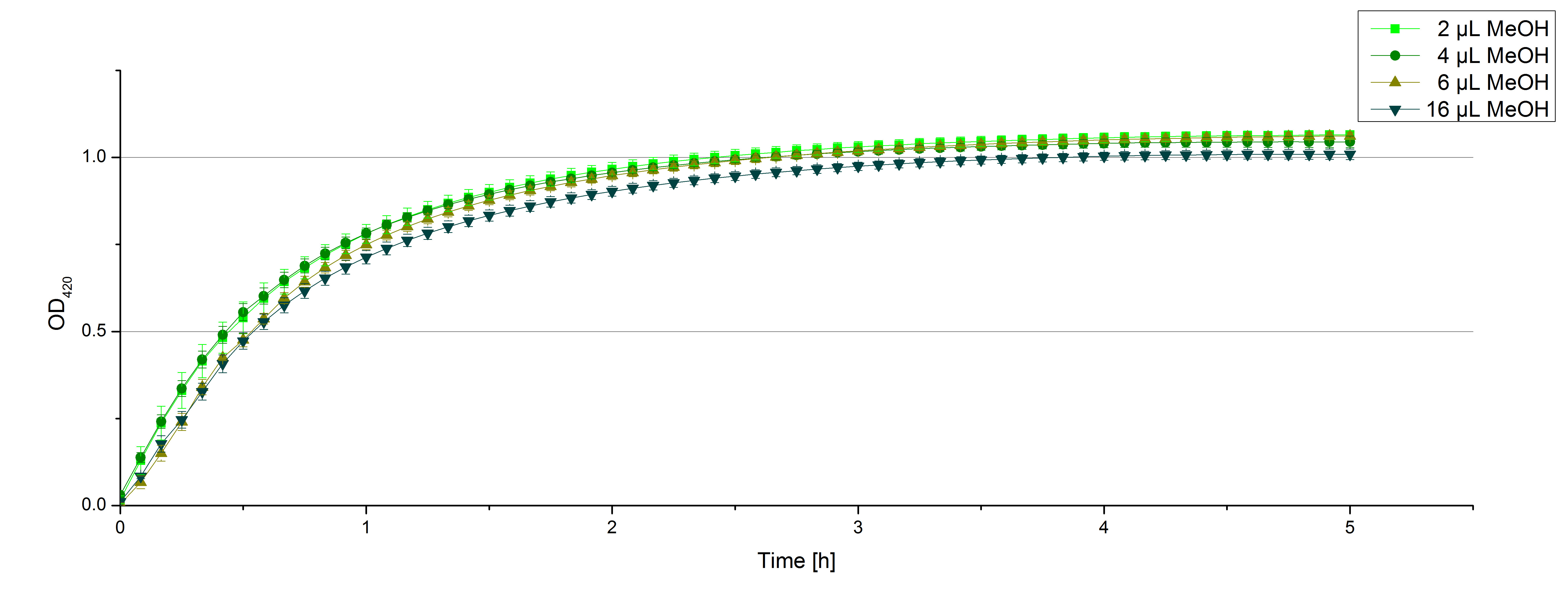
- After establishing our recipe for activity measurements we were curious about different ABTS concentrations and wanted to make sure we took the right one for our approach. With this in mind we did our activity measurement with 0.1 U [http://www.sigmaaldrich.com/catalog/product/sigma/53739?lang=de®ion=DE TVEL0 laccase], 100 mM sodium acetate buffer (pH 5), ad 200 µl H2O dest., but with two new ABTS concentrations, namely 0.2 mM and 0.05 mM ABTS (see Fig. 1). It turned out that using 0.05 mM ABTS leads to a low maximum of saturation but reaches it quickly. With 0.2 mM ABTS the opposite occurs: the activity curve is saturated at a OD420 of ~2.7 but it needs more time to reach its maximum. Knowing this we are happy using 0.1 mM ABTS because it is saturating slowly and the maximum is not too high. 0.1 mM ABTS is therefore established!
- After establishing our recipe for activity measurements we were curious about different ABTS concentrations and wanted to make sure we took the right one for our approach. With this in mind we did our activity measurement with 0.1 U [http://www.sigmaaldrich.com/catalog/product/sigma/53739?lang=de®ion=DE TVEL0 laccase], 100 mM sodium acetate buffer (pH 5), ad 200 µl H2O dest., but with two new ABTS concentrations, namely 0.2 mM and 0.05 mM ABTS (see Fig. 1). It turned out that using 0.05 mM ABTS leads to a low maximum of saturation but reaches it quickly. With 0.2 mM ABTS the opposite occurs: the activity curve is saturated at a OD420 of ~2.7 but it needs more time to reach its maximum. Knowing this we are happy using 0.1 mM ABTS because it is saturating slowly and the maximum is not too high. 0.1 mM ABTS is therefore established!
Wednesday May 30th
>
- Team Cloning Bacterial Laccases:
- After plasmid isolation we digested our plasmids with NotI to see if the colony PCR was correct and our laccases are in the backbone. The agarose gel showed that for all of the different plasmids we had at least one plasmid with two DNA bands on the correct height in agarose gel.
Thursday May 31st
- Team Cloning of Bacterial laccase:
- We sent the isolated pSB1C3 plasmids with xccl(T7)_His, Bpul(T7)_HIS and Ecol(T7)_His for sequencing.
Friday June 1st
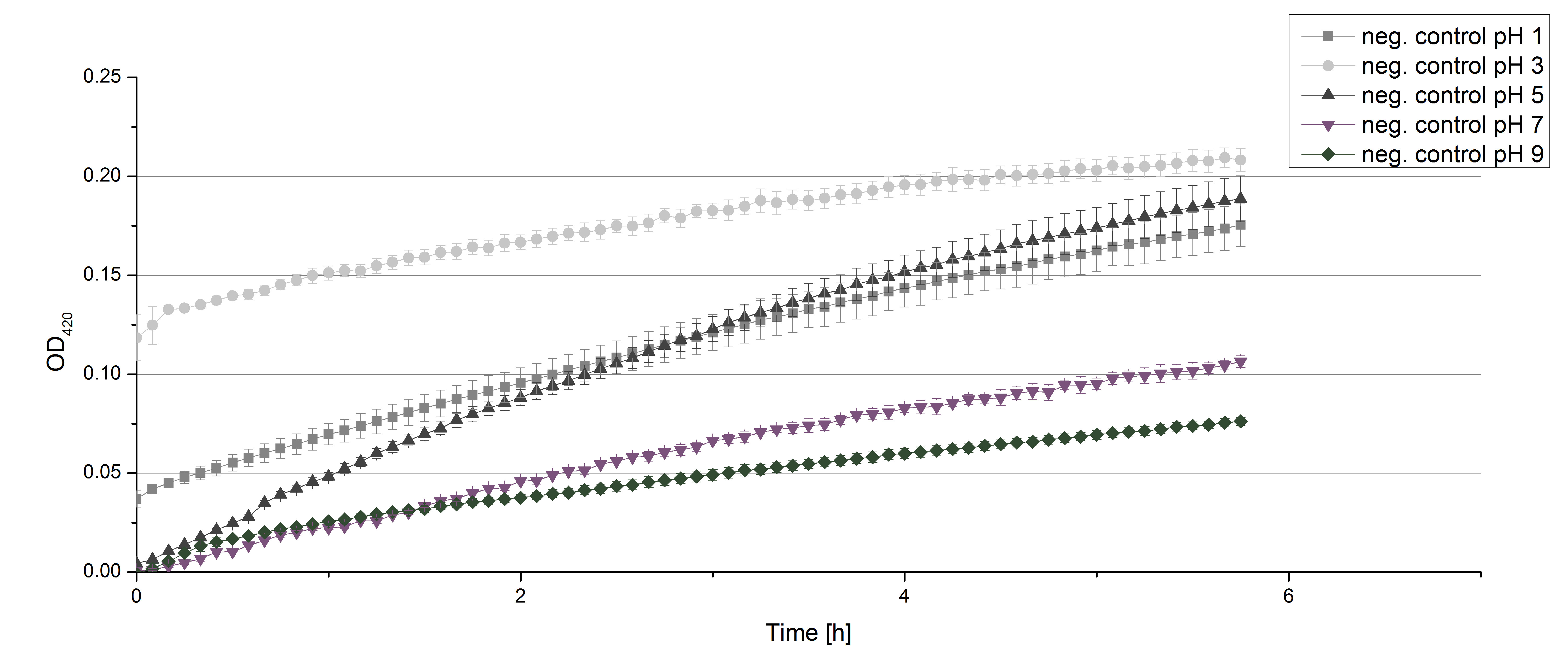
- Team Activity Test:
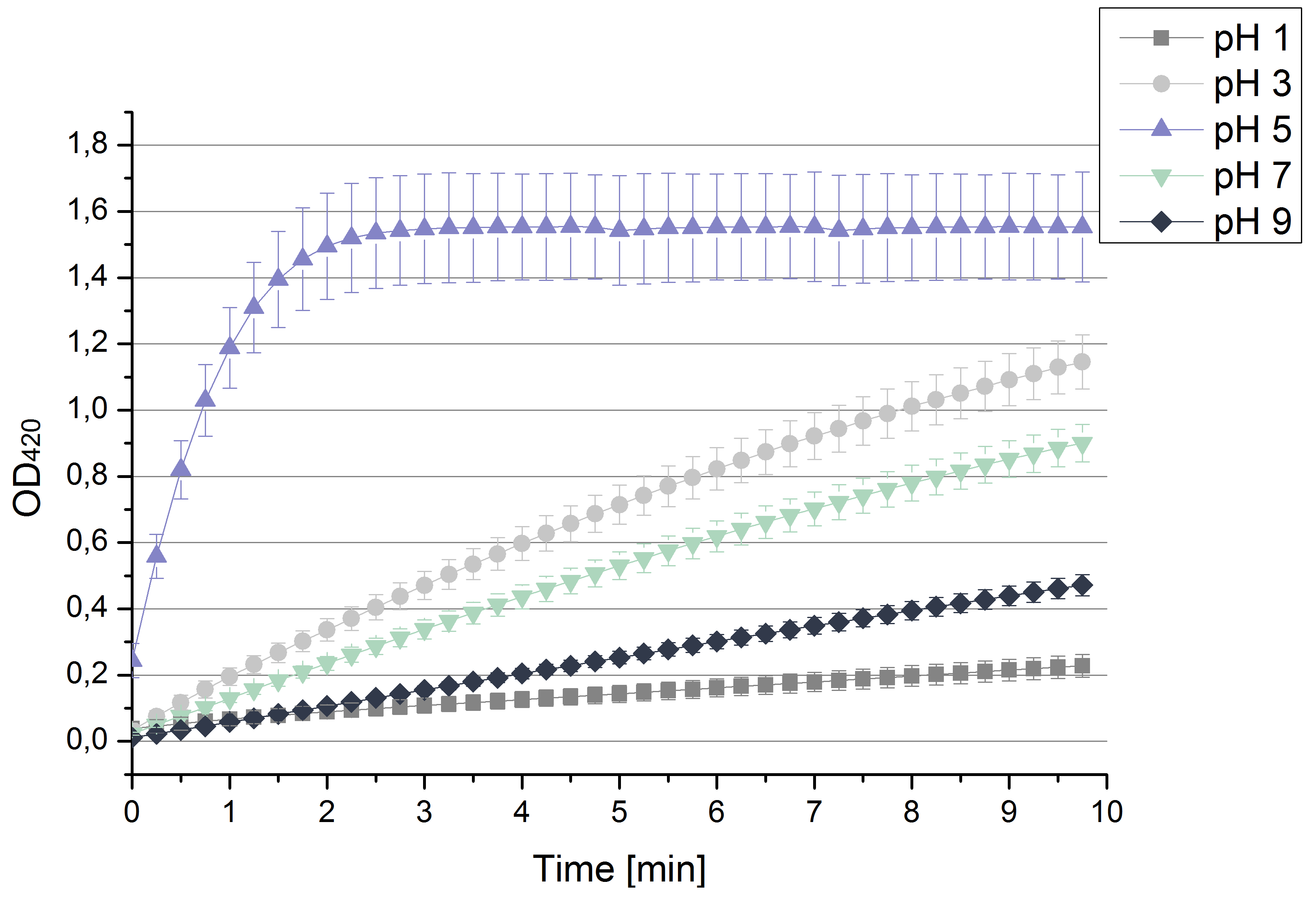
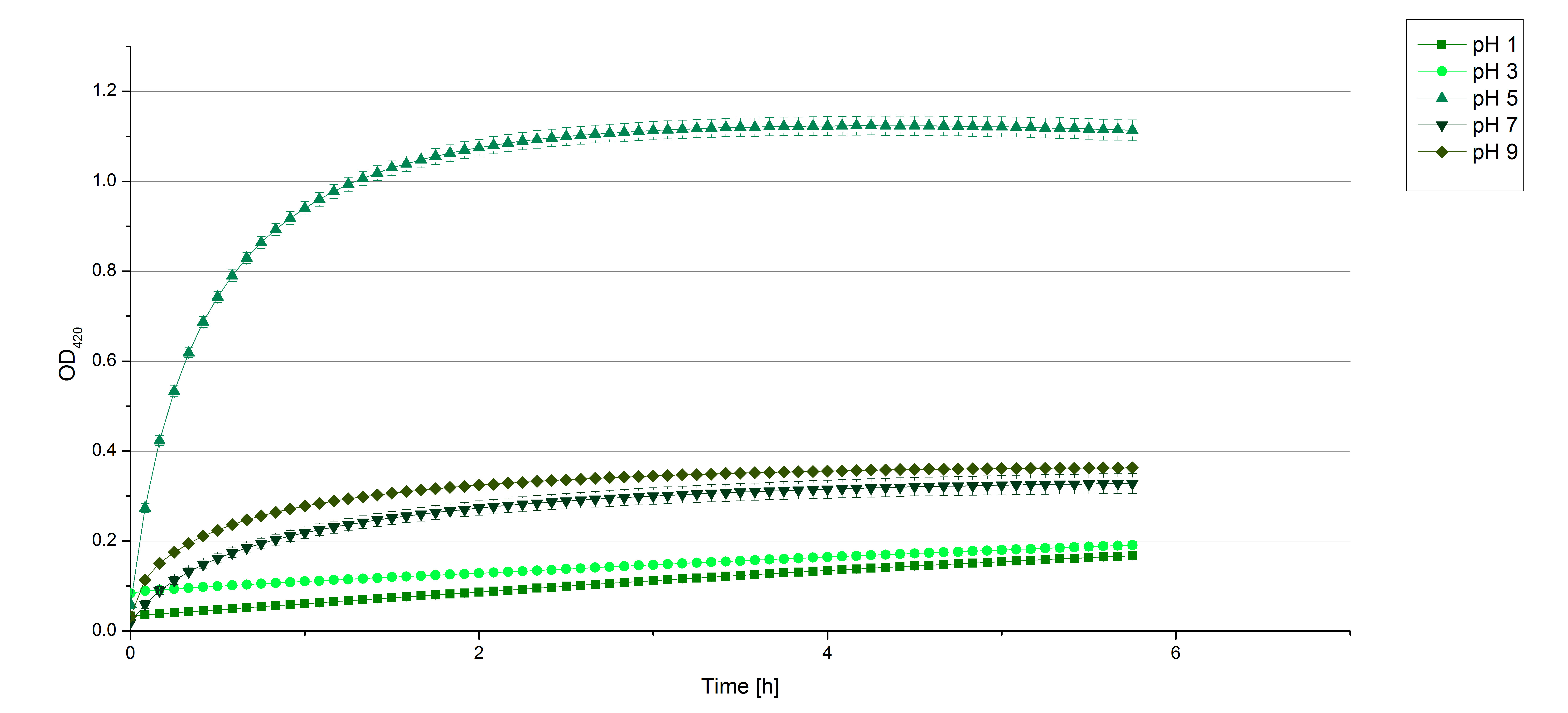
- Today we prepared for our next measurements by setting up the sodium acetate buffer in different pHs. We choose to test the activity of TVEL0 in a pH-Gradient of 1,2,5,7 and 9.
Saturday June 2nd
Sunday June 3rd
- Team Cloning of Bacterial Laccases:
- PCRs of genomic DNA on bhal from B. halodurans C-125 we ordered from [http://www.dsmz.de/ DSMZ] before. We handled the cells in the same way we did with T. thermophilus before and soluted the lyophlized cells in water and boiled them before PCR. After PCR we cleaned up the product with gel electrophoresis and PCR clean-up kit. However the DNA amount was so low that we had to do the PCRs again.
Summary of Week 6
hier eine Zusammenfassung
Week 6 (06/04 - 06/10/12)
Contents |
Weekly Seminar
- As part of the sponsoring with [http://www.merckgroup.com/en/index.html Merck] we will present our project in Darmstadt. Kevin, Nadine and Gabi travel to Darmstadt on 21th of june.
The team will gather on 14th, 15th and 18th of june to practice the presentation.
- Participants at the [http://www.cas.uni-muenchen.de/veranstaltungen/tag_synth_bio_2012/index.html CAS conference for Synthetic Biology]: Hakan, Derya, Miriam, Julia S., Gabi, Malak, Timo, Robert, Isabel, Nils.
- Robert is responsible for ordering the following BioBricks from the Partsregistry:
<partinfo>K500000</partinfo>
<partinfo>K500001</partinfo>
<partinfo>K500002</partinfo>
<partinfo>K500003</partinfo>
<partinfo>K392014</partinfo>
- The treaty with our sponsor [http://www.biocircle.com/en-ca/ BioCircle] is signed.
- All teams presented their lab results.
Monday June 4th
- Team Cloning of Bacterial Laccases:
- The Bacteria S. griseus and S. lavendulae has been delivered so we can start with PCRs. We set the first PCR with them as followed:
- PCR table
- The Bacteria S. griseus and S. lavendulae has been delivered so we can start with PCRs. We set the first PCR with them as followed:
| Material | Volume |
|---|---|
| Buffer (10x Phusion) | 10µL |
| Phusion Polymerase | 0,5µL |
| dNTPs | 1µL |
| Primer Mix | 1µL |
| Template DNA | 1µL |
| DMSO | 1,5µL |
| Water | 35µL |
- PCR program
| Temperature | Time |
|---|---|
| 1) 98°C | 7 mins |
| 2) 98°C | 20 sec |
| 3) 55°C | 20 sec |
| 4) 72°C | 1 min |
| 5) 72°C | 3 min |
| 6) 12°C |
Cycle between step 2 and 4 35 times.
- Team Fungal and Plant Laccases:
- Primerdesign for isolating a laccase from Arababidopsis thaliana cDNA. Since we want to express the laccase in E.coli we designed the primers like before for the bacterial laccases with T7 promotor and HIS-tag.
Tuesday June 5th
- Team Student Academy:
- Transformation of a plasmid mixture of either pMTE cp46 His and [http://partsregistry.org/Part:BBa_J04450 BBa_J04450] or pMTE cp46 His and [http://partsregistry.org/Part:BBa_J23100 BBa_J23100]. We plated both on LB agar without antibiotics and with Kanamycin. The first one was also plated on LB agar with ampicillin and the second on LB agar with chloramphenicol. Result: Works as expected. [http://partsregistry.org/Part:BBa_J23100 BBa_J23100] has a more intense fluorescence and was chosen for the experiment.
- Team Cloning of Bacterial Laccases:
- The sequencing results for isolated plasmids xccl(T7)_His, bpul(T7)_His and ecol(T7)_His came. The results showed that only the xccl(T7)_His was ok – our first finished biobrick *yeha*. We proudly name it <partinfo>BBa_K863015</partinfo>. The sequence of 'ecol(T7)_His showed that there are missing 4 bases in the promoter region and the bpul(T7)_His sequence showed a mutation which leads to another amino acid in protein sequence.
- Again we did PCRs on T. thermophilus laccase and B. halodurans laccase with B.halo_FW_T7 / B.halo_FW_HIS and T.thermo_LAC_FW_T7 / T.thermo_LAC_RV_HIS primers and purified the product,this time with enough material for a restriction.
- Team Activity Tests:
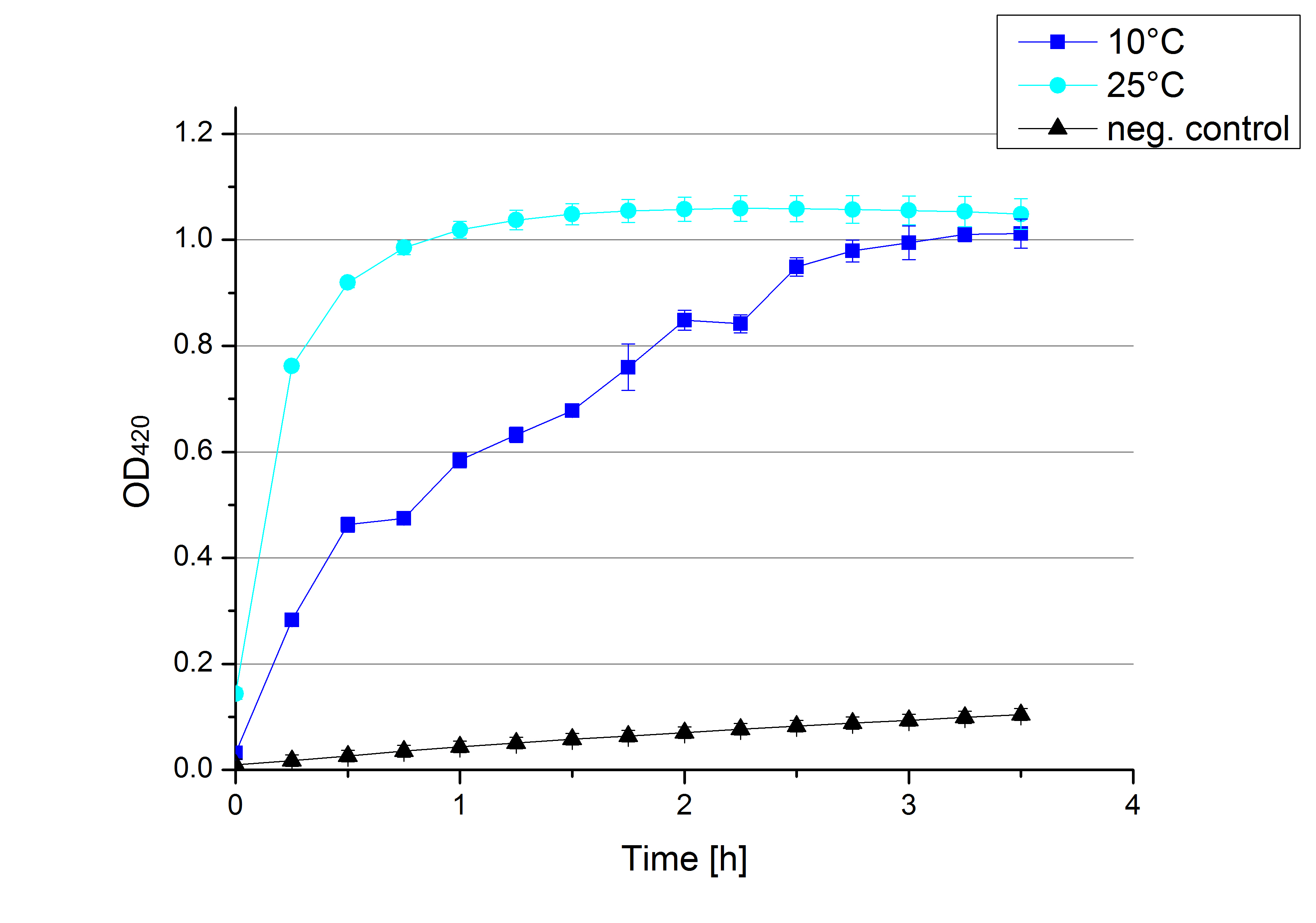
- After testing the T. versicolor laccase under conditions that are optimal (pH 5, 25°C ) according to the literature we now started further characterization under different pHs. We analyzed the laccases behavior when working in 100 mM sodium acetate buffer at pH 1, 3, 7 an 9. Result: We agree with the literature that pH 5 seems to make the laccase happy. Since not all waste waters (especially those here in Germany) are not as warm as 25°C we now wonder what our laccase might do when exposed to lower temperaturers. Stay tuned.
Wednesday June 6th
- Team Wiki: Yay for Team Wiki´s first entry. Our first steps with the iGEM Bielefeld 2012 Wiki contain thinking about contents, layouts, programming and responsibilities. Our first rules are:
- we are programming static pages in HTML and all the other pages (those that will be updated by all team members) in wiki code.
- we created all pages and will fill them up with some nice and beautiful content constantly from now on.
- Our temporary banner contains our outstanding logo and a DNA but we will set up a new layout soon.
- Team Cloning of Bacterial Laccases: Digest of tthl(T7)_His and bhal(T7)_His PCR products and ligation in pSB1C3 backbone. After that we transformed the plasmids in competent E. coli KRX cells.
Thursday June 7th
- Team Student Academy:
- Repeating of Transformation of 06/05 to verify the function. It is reproducible :)
- Team Cloning of Bacterial Laccases: Colony PCRs of the transformed colonies from yesterday's transformation showed some positive PCR bands.
- Team Modeling: becoming acquainted with matlab while reading the manual
Friday June 8th
- Team Cloning of Bacterial Laccases:
- We plated colonies for plasmid isolations on new plates and made a control restriction with NotI. The electrophoretic separation showed gel bands in the right height for the Tthl(T7)_His and with bhal(T7)_His.
Saturday June 9th
Sunday June 10th
Summary of Week 7
hier eine Zusammenfassung
Week 7 (06/11 - 06/17/12)
Contents |
Weekly Seminar
- Agatha and Saskia will perform a mRNA isolation from Arabidopsis thaliana.
- Everything is in progress. Check.
Monday June 11th
- Team Cloning of Bacterial Laccases:
- Prepared plasmids for sequencing. We sent another isolated plasmid with'ecol(T7)_His. Also tthl(t7)_His, bahl(T7)_His and bpul(T7)_His plasmids were ready for sequencing.
- Team Fungal and Plant Laccases:
- We looked for the sequences of the laccase of Trametes versicolor in different databases. By searching the specific sequences we realized, that we have to pay special attention to the strains the sequnces came from. During our search we cannot find any published strain in the DMSZ or other straincollections. So we decided to write e-mails to different working groups all over the world to get the published strains.
- Team Immobilization:
- Today, we took our first step: we started searching for publications which describe different methods for the immobilization of laccases, especially those from Trametes versicolor in order to figure out the best method that suits our laccases. Some of the papers are listed below:
- [http://www.ncbi.nlm.nih.gov/pubmed/22398306 Recent developments and applications of immobilized laccase.]
- [http://www.hindawi.com/journals/er/2011/725172/ Enzyme-Catalyzed Oxidation of 17β-Estradiol Using Immobilized Laccase from Trametes versicolor]
- [http://www.ncbi.nlm.nih.gov/pubmed/17917725 Reactive blue 19 decolouration by laccase immobilized on silica beads]
- [http://www.ncbi.nlm.nih.gov/pubmed/16988786 Alginate/carbon composite beads for laccase and glucose oxidase encapsulation: application in biofuel cell technology.]
- [http://www.sciencedirect.com/science/article/pii/S1381117709002045 A disposable Laccase–Tyrosinase based biosensor for amperometric detection of phenolic compounds in must and wine]
- Today, we took our first step: we started searching for publications which describe different methods for the immobilization of laccases, especially those from Trametes versicolor in order to figure out the best method that suits our laccases. Some of the papers are listed below:
Tuesday June 12th
- Team Student Academy:
- The whole experiment was tested by another team member to plan the course.
Wednesday June 13th
- Team Cloning of Bacterial Laccases: Since the GC amount of the S. griseus and S. lavendulae laccases are high we used betain to solve the PCR problem. Addition of betain did not change anything on the results, we still didn't got our laccase DNA.
- Team Modeling: Programming our first differential equation and finding the ODE15s function witch solves these equations.
Thursday June 14th
- Team Cloning of Bacterial Laccases:
- Because our PCRs have not worked well we thought it may depends on the primer annealing temperature so we did gradient PCR with the same conditions as before (PCR June 4th). But this also showed no result. Because we made Coloyn PCRs from the arrived DSMZ reaction tubes our next idea was to cultivate the bacteria in media and isolate genomic DNA.
- Team Activity Tests:
- Since our Tecan microplate reader is not able to actively cool down to 4 °C we got the chance to meet the photometer Carry. Check "protocols" for further information about her. We used the same set up with 100 mM natrium acetate buffer, 0,1 U T. versicolor laccase and 0,1 mM ABTS as before but now measured at 4°C. Our team is planning to visit a municipal sewage plant for getting some insights into the water conditions there, so we will for sure test other temperatures after having more information. Let´s hope the water there is a little warmer since laccase does not seem to be totally satisfied at 4°C. I would not either.
Friday June 15th
- Team Cloning of Bacterial Laccases:
- Sequencing of the pSB1C3 plasmid with bhal(T7)_His was ok. In conflict to our reference sequence there was a point mutation in the DNA sequence but this mutation doesn’t lead to another amino acid. So..next BioBrick (<partinfo>BBa_K863020</partinfo>) is ready to use!
- The sequenced plasmid bpul(T7)_His showed again the same mutation in the laccase ORF compared to the reference sequence. We concluded that probably the PCR amplification caused the point mutation. So we did the digest of bpul(T7)_His PCR products from a new PCR, ligated it in pSB1C3 backbone and transformed it in competent KRX cells. Additionally we did the digest tthl(T7)_His and the ligation in pSB1C3 backbone again.
- Team Fungal and Plant Laccases:
- The whole week we were looking for different sources of the laccase sequences and the corresponding strain, we find an interessting study of the Federal Environmental Agency (Umwelt-Bundes-Amt Germany). In this study they analysed the oppurtunities to synthesis different chemical compounds with the aid of different laccase of Tramtetes versicolor. In this document we found a working group from the Institute of Biochemistry, Dept. of Biotechnology & Enzyme Catalysis at Ernst-Moritz-Arndt-University in Greifswald (Germany) who have isolated four sequences of different laccases of Trametes versicolor and the sequence of one Pycnoporus cinnabarinus. We have send a request for the plasmids containing cDNA sequences of five different laccases from Trametes versicolor and Pycnoporus cinnabarinus .
- Team Database: We finished our first tabel, you can add a new entry, delete it and edit it.
Saturday June 16th
Sunday June 17th
Summary of Week 8
hier eine Zusammenfassung
Week 8 (06/18 - 06/24/12)
Contents |
Weekly Seminar
- Julia V. presents the database to have an easier overview over the labwork, the biobricks and as a digital lab diary.
- the last changes have been implemented into the press release, it is about time to let the world know about our project.
- Everyone please keep the iGEM deadlines in mind.
- Our advisor Timo presents: iGEM 101: Introduction to the wiki.
- Julia S is presenting her progesses in designing the Pichia pastoris shuttle vector.
- Modelling: Julia V. is thinking about how expression rates and promoter activity can be implemented into the model. Sebastian tries to model a sewage treatment plant.
Monday June 18th
- Team Cloning of Bacterial Laccases:
- We started Colony PCRs on the colonies from June 15th transformation and picked positive colonies to plate them for plasmid isolation. Sadly we just had positive clones for tthl(T7)_His and not for bpul(T7)_His.
- Team Site Directed Mutagenesis:
- Reviewed all Assembly-Standards and made a list of all illegal restriction-sites:
- EcoRI, NotI, PstI, SpeI, XbaI (Silver), AgeI, NgoMIV (Freiburg), BamHI, BglII, XhoI (Berkeley)
- Decided to not care for restriction-sites of the Berkeley-assembly (even if it is a great assembly for protein-fusion), because the used Vector (pSB1C3) already has two XhoI-restriction-sites
- Team Fungal and Plant Laccases
- We have not received any reply to our e-mail. Team meeting to discuss the further way forward. We decided to send a second wave of e-mails to the different working groups, try to call the working groups(if possible) and to look for strains of the corresponding and published sequences in additional straincollections.
Tuesday June 19th
- Team Wiki:
- While using the lab journal more frequently there came up some questions.
- How detailed do we plan to write our lab journal entries?
- Do we want to write in keywords or explain everything in full sentences?
- Do we want to note every little detail about every successful or unsuccessful experiment or just the main important aspect?
- We discussed, sighted some former iGEM team wikis and decided:
- each team is responsible for their own lab journal entries
- we divide our lab journal in weeks and days to prevent it from looking too chaotic.
- the texts are supposed to state which team is writing, which experiment has been done and what the main aspects were. Also we will write about successful experiments, as well as problems and solutions we came up with. If possible links to protocols with further information shall be created.
Wednesday June 20th
- Team Site Directed Mutagenesis:
- Imported sequences of most of the used bacterial Laccases into Clonemanager and analysed their restriction-sites:
- bhal has no illegal restriction-sites
- ecol has one NgoMIV-restriction-site
- bpul has one XbaI-Restriction-site and one mutation two AgeI- and two NgoMIV-restriction-sites (Decided to delete the Freiburg-restriction-sites would take too much time)
- tthl has one PstI-restriction-site and two NgoMIV-restriction-sites (Decided to delete the Freiburg-restriction-sites would take too much time)
- xccl has two PstI, one AgeI and seven NgoMIV-restriction-sites (Decided not to change the NgoMIV-sites, since to mutate seven would take too much time)
- Team Modeling:
- Finding out, that the "normal" Michaelis-Menten kinetic isn't the right kinetic to model our situation, because therefor you need a high and steady state concentration of the substrates. We have low concentrations and not really study state. We found a transformed equation.
Thursday June 21st
- Team Cloning of Bacterial Laccases:
- Plasmid isolation and control digest with NotI on tthl(T7)_His and luckily this time the bands were where they should be. Again and again we did transformation of ligation with bpul(T7)_His laccase in pSB1C3 backbone..all fingers are crossed that this time we have colonies with the correct plasmid.
- Team Activity Tests and Team Immobilization:
- After all this characterizing we feel so much closer to our T. versicolor laccase that its about time to make some activity test under immobilized conditions. So now we are cooperating with Team Immobilization. We have thought about many ways how to immobilize the laccase and decided to give Silica Beads the first try. Check the Immobilization Team´s protocol for further information. Our main problem was how to measure the samples with all those beads in it. Tecan will probably be confused and give us some false values due to the beads that are disturbing its laser. So we need a way to get the beads out (and thus also stop the reaction) at a very precise point of time. Centrifugation wasn´t an option because it would simply take too long and not stop the reaction exactly in the second we want. While checking the internet for solutions we found Multi-Well Membrane-Bottom Filter Plates. Those are supposed to work in a similar way then our regular plates which we used for the Tecan but furthermore those plates contain a membrane that sieve the liquids through the filter when centrifugated. Thus the beads are separated and the ABTS-Buffer solution can me analyzed at 420 nm for oxidized ABTS. The plates will need a while before they arrive here at the CeBiTec, so we decided to first find out what the optimal amount of beads is and whether the beads might also bind ABTS (see lab journal Team Immobilization).
- Team Immobilization:
- So after a lot of reading and discussion, we decided to try immobilization using beads. Since silica beads were already available in our lab (from last year’s team), we decided to give them a try. The first challenge was to find out a convenient ratio of beads/laccase. According to the protocol of last year’s team, the ratio 1:1000 was used (1000mg beads/ 1 mg protein). Therefore we decided to try the ratios 1:500, 1:1000 and 1:1500. We prepared different buffers: HBSS buffer, Recrystallization buffer both of which were used with silica beads; in addition to Britton-Robinson Buffer, which was mentioned in publications as the best buffer for laccase immobilization. Laccases from TVEL0 were incubated with silica beads and different buffers at room temperature for 4hours on a rotator. After that, we collected the supernatants and delivered them to the team “Activity test” and waited for the good news.
Friday June 22nd
- Team Cloning of Bacterial Laccases:
- Because our PCR didn't work on the boiled lyophilized cells we used [http://www.carlroth.com/catalogue/catalogue.do;jsessionid=2E5F0AF60BF1F28909D8475CF0053386?favOid=00000000000180e500070023&act=showBookmark&lang=de-de&market=DE CASO Medium] for cultivation of S. griseus and S. lavendulae.
- Team Immobilization:
- Unfortunately, the activity test results weren’t promising. According to publications, immobilization via covalent binding is the most widely used method. Silica dioxide bead offer only an adsorption of laccases. Therefore, we decided to order CPC-(controlled pore carrier) silica beads, to which laccases covalently bind, especially that we found some papers with protocols and activity tests proving their efficiency.
Saturday June 23rd
- Team Cloning of Bacterial Laccases:
- The cultured S. griseus and S. lavendulae bacterials has been centrifuged at 13.000 rpm for 5 minutes. After this step we ribolyzed the pellet in 1 ml TE-Puffer and set a PCR reaction after. But we still haven't had any results.
- Colony PCRs on the transformations with plasmid with bpul(T7)_His and plating positive colonies.
- Team Database:
- In the last wekk we finished the tabels Eppi, User, Sequencing and BioBrick.
- We have a problem with the search function, we use the false SQL statement and allready deleted entries are shown.
- In the next week we have to write a new statement.
Sunday June 24th
- Team Cloning of Bacterial Laccases:
- We isolated plasmids and did control digests with NotI. We finally had a positive restriction digest for bupl(T7)_His. So we prepared this plasmids and the plasmid tthl(T7)_His which we isolated some days before for sequencing.
Summary of Week 9
hier eine Zusammenfassung
Week 9 (06/25 - 07/01/12)
Contents |
Weekly Seminar
- evaluation of the presentation, we did in Darmstadt as part of our sponsoring through [http://www.merckgroup.com/en/index.html Merck]
- Merck is strongly interested in our results, because the company has problems with lignin in the waste water (and laccases can be used for degradation of lignin, too).
- We were advised to use silica-beads, pretreated with carbodiimide, to immoblize our enzymes
- We decided to visit at least one sewage treatment plant as part of our Human Practices and we will try to integrate the treatment plants into our modelling.
- Everything is prepared for our summer school. Kevin and Gabi will present the experiment and
Nadine, Kevin, Gabi, Mo, Miriam, Derya and Isabel will advise the pupils.
- Gabi, Timo and Robert went to the strategy process [http://www.biotechnologie2020plus.de/BIO2020/Navigation/DE/root,did=152728.html "Biotechnologie2020+"] in Berlin.
Monday June 25th
- Team Cloning of Bacterial Laccases:
- Retried the DNA isolation from S. griseus and S. lavendulae without any success.
- Team Fungal and Plant Laccases:
- Phonecall with the Leader of the working group of the [http://biotech.uni-greifswald.de/ University Greifswald Prof. Dr. U. Bornscheuer]. We explained our project and asked, if we can get the sequnces of the Trametes versicolor laccases. We got the commitment for getting the laccase sequences and plasmids containing the sequences (four laccases of Trametes versicolor and one of Pycnoporus cinnabarinus).
Tuesday June 26th
- Team Fungal and Plant Laccases:
- The thing about plants is that they have to grow. Fortunately we got 6 beautiful 4 weeks-old wildtype plants from Patrick Treffon from the Institute of Plant Physiology and Biochemistry at Bielefeld University. With the help of the [http://bar.utoronto.ca/efp/cgi-bin/efpWeb.cgi efp-Browser] we found out that the [http://www.ncbi.nlm.nih.gov/protein/AAM77221.1 laccase] in A. thaliana is only expressed in the developing seeds. So we now have to wait for the siliques to develop.
- Team Shuttle Vector:
- Prepare the YPD Media for cultivation of the yeast strains Pichia patoris X-33 (wild type) and GS115 (Invitrogen). Both organisms are provided from the [http://www.techfak.uni-bielefeld.de/ags/fermtech/ chair of Fermentation Engineering] (D5) from Dr. Thomas Hug.
Wednesday June 27th
- Team Shuttle Vector:
- Cultivation of Pichia pastoris X-33 and GS115 in YPD media for isolation of the genomic DNA.
- Team Activity Tests:
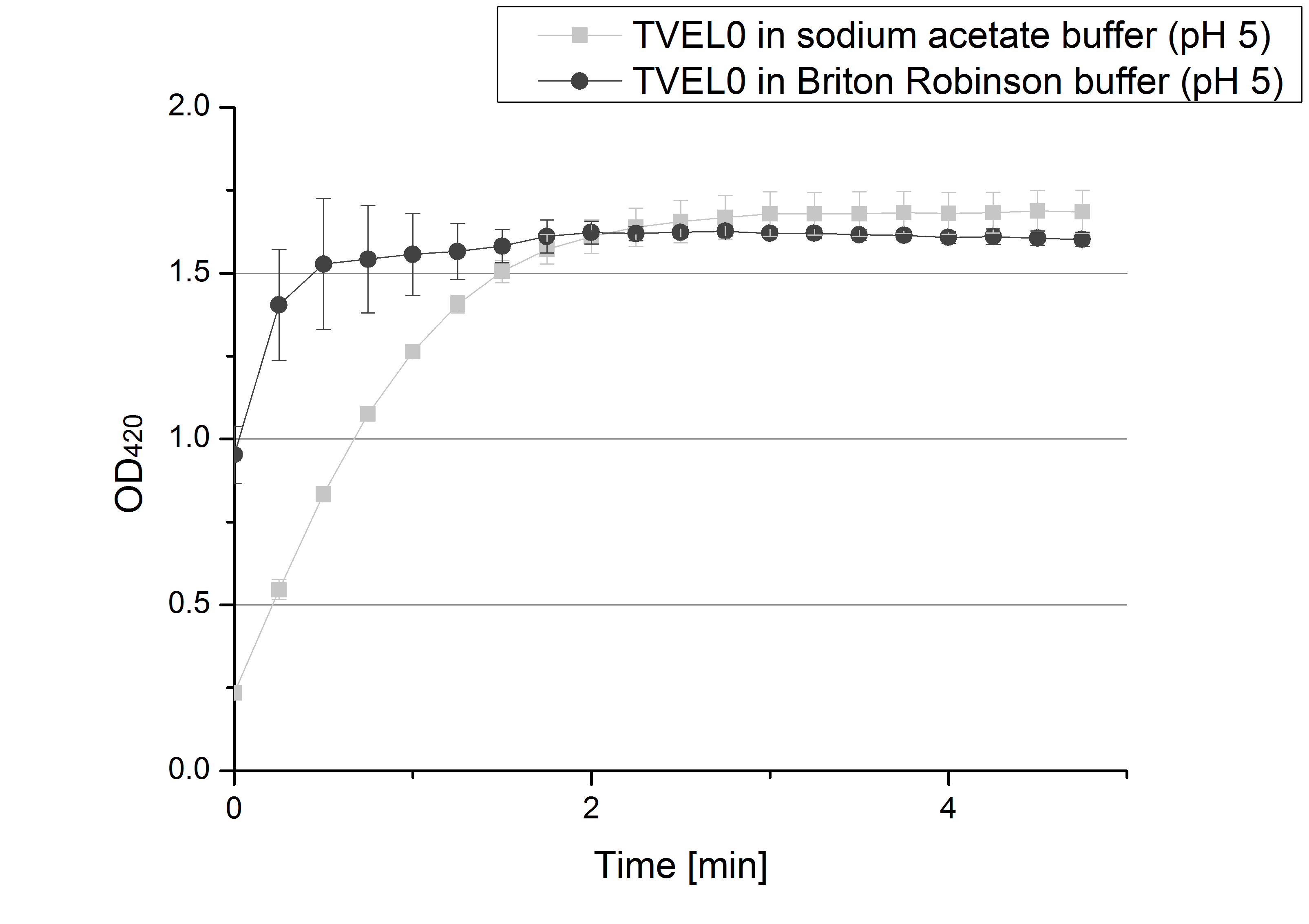
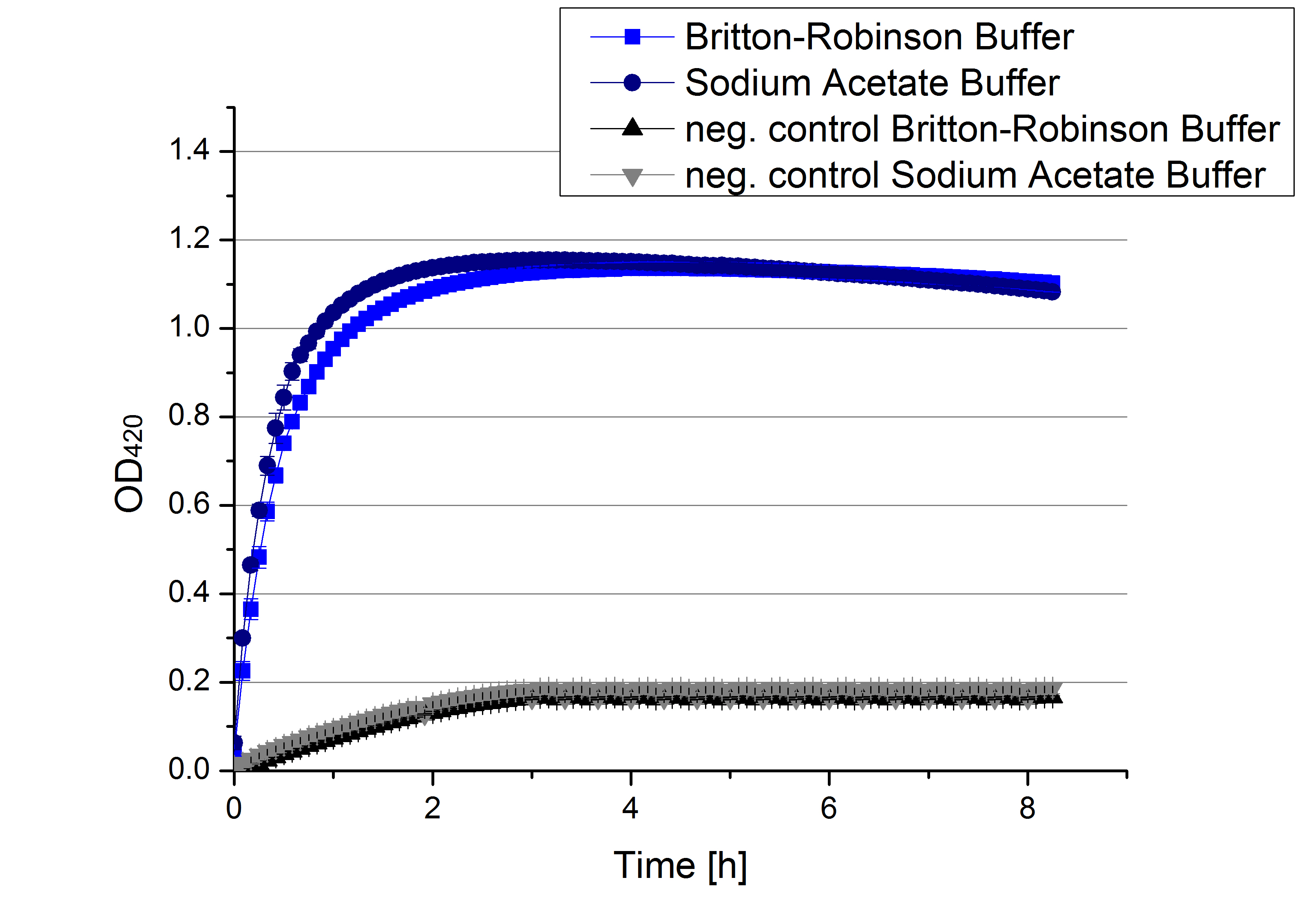
- We like our new cooperation with Team Immobilization. The thing is, that they don´t like our buffer. Sodium acetate (pH 5) seems perfect for activity tests but apparently not for immobilization. What they prefer is a Britton-Robinson buffer (pH 5). To find out whether there is a difference between the two buffers that causes different activity habits of our laccase TVEL0, we setup comparable measurements with the two buffers and TVEL0. We concluded that the laccase in sodium acetate buffer shows a slower saturation but all in all both laccase samples reach the same maximum so that it is ok for us to use both buffer systems.
Thursday June 28th
- Team Wiki:
- Today we browsed our wiki and were not very impressed: it's a lonesome place. So we started to think of how we could blow a little more life into it. For this, texts should appear soon on our wiki. To manage this bunch of work, we divided the subtopics of our wiki and appointed them to group members. Now everyone has a topic which he is responsible for. And that includes writing the texts, uploading pictures and keeping the represented information updated. Before anybody had the chance to disappear behind his/her notebook being busy editing his own page, we had to establish our wiki rules:
- Use the standardized formatting as presented in our example page.
- Try to edit your text without using HTML code as far as possible and use wiki code instead. Useful advices when using wiki code are represented on our example page, too.
- If you want to change anything that does not belong to your scope of duties, ask kindly the person of charge and make sure he/she is fine with it.
- Make your text more understandable by using images and charts. But remember: you are only allowed to upload pictures if you own them or if they are published without licenses.
- Team Shuttle Vector:
- Isolation of genomic DNA from Pichia pastoris GS115 (Invitrogen) and the wild type X-33 with the Wizward genomic DNA purification system Kit (Promega).
- Phusion PCR was done with the following primers to check that we created the right primers.
| Used primers | Template | successful amplification |
|---|---|---|
| 5'AOX_FW and 5'AOX_RV | pPICzA | yes |
| 3'AOX_FW and 3'AOX_RV | pPICzA | no |
| pAOX1_FW and pAOX1_RV | pPICzA | yes |
| tAOX1_FW and tAOX1_RV | X-33 | yes |
Friday June 29th
- Team Cloning of Bacterial Laccases:
- Sequencing results showed that even with a new PCR product the same mutation occurs in bpul(T7)_His so it is probably already present on the plasmid which was sent to us. So we decided to use this plasmid. We give him the name <partinfo>BBa_K863000</partinfo>. Tth(T7)_His showed a positive sequencing result, too. So we have <partinfo>BBa_K863010</partinfo> ready for use!
- Team Shuttle Vector:
- Primer design was done for the shuttle vector. The following primers create fragments with a 5' overlap. Were ordered the primers: pSB1C3-5aox1-f, pSB1C3-5aox1-r, 5aox1-mfalpha1-f, 5aox1-mfalpha1-r, mfalpha1-aarI-taox1-f, mfalpha1-aarI-taox1-r, taox1-phis4-f, taox1-phis4-r, phis4-kozak-his4-f, phis4-kozak-his4-r, his4-3aox1-f, his4-3aox1-r, 3aox1-pSB1C3-f , 3aox1-pSB1C3-r.
Saturday June 30th
- Team Student Academy:
- We prepared a script for pupils containing background information and a protocol and wrote an abstract for the school academy program.
Sunday July 1st
Summary of Week 10
hier eine Zusammenfassung
Week 10 (07/02 - 07/08/12)
Contents |
Weekly Seminar
- Summer school: we will organize a barbecue and drinks, therefore some stuff has to be bought.
- We now have contact to the first sewage treatment plant.
- Malak will translate our abstract into english for the [http://www.cas.uni-muenchen.de/veranstaltungen/tag_synth_bio_2012/index.html CAS conference] in munich.
Monday July 2nd
- Team Student Academy:
- During the whole week the two presentations for school academy were prepared and the barbecue for the pupils was organized. In the lab the last preparations were made.
- Team Fungal and Plant Laccases:
- Sending a request for plasmids containing cDNA sequences of five different laccases from Trametes versicolor and Pycnoporus cinnabarinus from the Institute of Biochemistry, Dept. of Biotechnology & Enzyme Catalysis at Ernst-Moritz-Arndt-University in Greifswald.
Tuesday July 3rd
- Team Site Directed Mutagenesis:
- Decided how to insert silent mutations to get rid of the restriction-sites with an eye on the codon-usage of host- and scource-oragnism, using [http://www.kazusa.or.jp/codon/ the Kazusa “Codon Usage Database”]
- ecol’s illegal NgoMIV will be deleted by changing ggG to ggA (Glycin) at 2307
- tthl’s illegal PstI will be deleted by changing caG to caA (Glutamine) at 2796
- bpul’s illegal XbaI will be deleted by changing ctA to ctT (Leucin) at 2883
- bpul’s mutation will be deleted by changing Gag (Glutamat) to Aag (Lysin) at 2317
- xccl’s first illegal PstI will be deleted by changing ctG to ctC (Valine) at 2247
- xccl’s second illegal PstI will be deleted by changing ctG to ctC (Valine) at 3633
Wednesday July 4th
- Team Student Academy:
- Final meeting with all instructors of the student academy to talk about all details.
- Team Cloning of Bacterial Laccases:
- After the failed attempts to isolate the laccase genes from Streptomyces lavendulae and Streptomyces griseus we assumed that maybe the sequences from this laccases and the laccase sequences we designed the primers for were to different to get a product (look here for more details). So we used the sequences of the laccase genes we tried to isolate from the Streptomycetes strains and blasted them against an intern database from our university. The results showed that we could use genomic DNA from Streptomyces sp. tuebingen, S.roseochromogenes and Streptomyces sp. goettingen, from which we can get already isolated genomic DNA. Therefore we designed primers for the isolation of the predicted laccase genes from these three strains.
- Team Shuttle Vector:
- Phusion PCR was done with the following primers to see if the genomic DNA can used as template for the part 3OAX1. For creating the mating factor alpha 1 [http://partsregistry.org/Part:BBa_K863206 K863206] the plasmid pPICzA was used, because we did not have the wildtype strain of Saccharomyces cereviceae. For protein secretion we used the mating factor alpha 1 from Sacharomyces sereviceae because it secrete protein in the outer media better than the secretion factor from P. pastoris (LITERATURANGABE)
| Used primers | Template | Successful amplification | For BioBrick part |
|---|---|---|---|
| 3'AOX_FW and 3'AOX_RV | X-33 | yes | [http://partsregistry.org/Part:BBa_K863201 K863201] |
| 3'AOX_FW and 3'AOX_RV | GS115 | yes | [http://partsregistry.org/Part:BBa_K863201 K863201] |
| Ko_alpha_FW and Ko_alpha_RV | pPICzA | yes | [http://partsregistry.org/Part:BBa_K863206 K863206] |
| tAOX1_FW and tAOX1_RV | X-33 | yes | [http://partsregistry.org/Part:BBa_K863203 K863203] |
Thursday July 5th
- Team Shuttle Vector:
- Some fragments for Gibson Assembly with overlapping DNA sequences were amplified via Phusion-PCR. The fragment 5'AOX1 was amplified with the primer pair pSB1C3-5aox1-f and pSB1C3-5aox1-r. The fragment MF-alpha1 was amplified with the primer pair 5aox1-mfalpha1-f and 5aox1-mfalpha1-r. The fragment 3'AOX1 was amplified with the primer pair his4-3aox1-f and his4-3aox1-r. The fragment pSB1C3 was amplified with the primer pair 3aox1-pSB1C3-f and 3aox1-pSB1C3-r.
- Team Shuttle Vector:
- Phusion PCR was done with the following primers to create fragments with 5' overlap for the Gibson assembly. Also a gel purification was done.
| Fragment | Used primers | Template | Successful amplification |
|---|---|---|---|
| 5AOX1 | pSB1C3-5aox1-f and pSB1C3-5aox1-r | X-33 | yes |
| Kozak + MFalpha1 | 5aox1-mfalpha1-f and 5aox1-mfalpha1-r | X-33 | yes |
| 3AOX1 | his4-3aox1-f and his4-3aox1-r | X-33 | yes |
| pSB1C3 | 3aox1-pSB1C3-f and 3aox1-pSB1C3-r | RFP::pSB1C3 | yes |
Friday July 6th
- Team Database:
- FINISHED!! We finished our database. The search statement is now correct. And here it is:
Saturday July 7th
- Team Fungal and Plant Laccases:
- We received an e-mail from the working group leader Prof. Dr. Bornscheuer (Institute of Biochemistry, Dept. of Biotechnology & Enzyme Catalysis at Ernst-Moritz-Arndt-University in Greifswald (Germany)). He send us a confirmation that the plasmids with the cDNA of four Trametes versicolor-laccases and one laccase of Pycnoporus cinnabarinus were sended to the iGEM Team Bielefeld.
Sunday July 8th
Summary of Week 11
hier eine Zusammenfassung
Week 11 (07/09 - 07/15/12)
Contents |
Monday July 9th
- Team Student Academy:
- This week the Student Academy took place. Today we held a presentation about the background of our experiments and answered all the questions the pupils had. Furthermore two of us participated at the an unconstrained meeting between the instructors and the pupils in the evening.
- Team Shuttle Vector:
- Phusion PCR was done with the following primers to create fragments with 5' overlap for Gibson assembly. Also a gel purification was done.
| Fragment | Used primers | Template | Successful amplification |
|---|---|---|---|
| his4 promoter | taox1-phis4-f and taox1-phis4-r | X-33 | no |
| Kozak + his4 | phis4-kozak-his4-f and phis4-kozak-his4-r | X-33 | no |
- Because no PCR fragments could be detected. The same PCR was running with 10 µL of 5M Betain in a sample of 50 µL and provide to the results, that no PCR fragment for the fragment pHIS4 could. But the fragment Kozak + his4 with an 5' overlap could be detected.
- Team Site Directed Mutagenesis: Council with Katharina Thiedig, who did the Site Directed Mutagenesis for the last years iGEM-Team-Bielefeld, about how to use the “QuikChange Primer Design”-program and the SDM-protocol
- Team Cellulose Binding Domain:
- Talked to Dominik Cholewa from the fermentation group of Bielfeld University about cellulose binding domains and got confirmed that they have some on plasmid and want to share them with us.
Tuesday July 10th
- Team Shuttle Vector:
Surprisingly, why the PCR for the fragment his4 promoter does not raise the theoretical fragment with X33 as template, we check the DNA sequence we used for primer design. For design the primer taox1-phis4-f and taox1-phis4-r we used the DNA sequence from the BioBrick [http://partsregistry.org/Part:BBa_K563000 BBa_K563000]. But this part was designed on DNA from Sacharomyces cereviceae. So we thought that we can not use this DNA sequence in Pichia pastoris. Our concerns were that the promoter site could not be detected by the transcriptional regulatory machinery. We tried to identified the promoter region upstream the his4 gene on the plasmid pPIC9K from Invitrogen. Some blasts later, we saw that the his4 ORF from this plasmid is virtually the same as the CDS in [http://www.ncbi.nlm.nih.gov/nuccore/537483 NCBI]. Therefore, to create primers we used the DNA sequence from the plasmid pPIC9K and used the genomic DNA from the wild type P. pastoris X-33 or PCR amplification. Thus the primers taox-his4-f, taox-his4-r and his4-3aox1-f02 were new designed and ordered.
- Team Site Directed Mutagenesis: Generated primer-sequences with Agilent Technologies “QuikChange Primer Design” for bpul, xccl, ecol and tthl and named the primers by their source-organism and place of mutation flanked on the left side by the original base and on the right by the mutated base.
- ecol-g2307a
- tthl-g2796a
- bpul-a2883t
- bpul-g2317t
- xccl-g2247c
- xccl-g3633c
- Team Cellulose Binding Domain: Searched NCBI for Cellulose, Chitin & Keratin binding motifs in accessible organisms; found two Chitin-binding-domains in Bacillus halodurans
- Team Student Academy:
- The experiments were performed by one half of the pupils in groups of 2-3. Afterwards we held a presentation about the iGEM competition, the project of the last two teams from Bielefeld and about our project. In the evening we had a barbecue with the pupils and the iGEM team with a lot of interesting discussions.
- Team Fungal and Plant Laccases:
- We got the sequences for five different fungal laccases from the requested plasmids from Monday 2nd and they wanted to send us plasmids which contain the cDNA sequences of five different laccases from Trametes versicolor and Pycnoporus cinnabarinus.
- tvel5 from Trametes versicolor
- tvel10 from Trametes versicolor
- tvel13 from Trametes versicolor
- tvel20 from Trametes versicolor
- pcil35 from Pycnoporus cinnabarinus
- We designed primer pairs with prefix and suffix overhanging ends for cloning in pSB1C3 and a primer pair for cloning in shuttle vector.
- The first primer pairs were designed with standard prefix and suffix sequence and 20 bases complementary to the start and end of the ORF sequences.
- Additional primer pairs were designed with AarI restriction site and Kozak consensus sequence before the first 20 bases from the start of the ORF (forward primers). The reverse primers were designed with the last 20 bases of the laccase genes and a terminator overhanging end and a AarI restriction site.
- We got the sequences for five different fungal laccases from the requested plasmids from Monday 2nd and they wanted to send us plasmids which contain the cDNA sequences of five different laccases from Trametes versicolor and Pycnoporus cinnabarinus.
Wednesday July 11th
- Team Cellulose Binding Domain: Looked the sequence of [http://www.ncbi.nlm.nih.gov/nuccore/M73817 Clostridium cellulovorans cellulose binding protein gene (cbp A)] up and made a Clonemanager-file. We used a protein-BLAST of the translated protein-sequence to find the location of the cellulose binding domain within the protein: We found it should be from base 103 to base 378 of the open reading frame.
- Team Student Academy: The experiments were performed by the second half of the pupils and the groups from yesterday analyzed their plates.
- Team Cloning of Bacterial Laccases: We made glycerin cultures of the finished BioBricks.
- Team Shuttle Vector: The gel purified fragments 3AOX1 ([http://partsregistry.org/Part:BBa_K863200 K863200]), MFalpha1 ([http://partsregistry.org/Part:BBa_K863206 K863206]), pAOX1, tAOX1 ([http://partsregistry.org/Part:BBa_K863203 K863203]) and the plasmid pSB1C3 were double digested with EcoRI and SpeI as in the follow. Mix 10 µL of PCR amplification and pSB1C3, respectively to 18 µL bidest. water, add2 µL of 10x buffer O, 0,5 µL of each enzyme. After incubation for 1 h at 37 °C and inactivation at 80 °C for 20 min the plasmid was dephosphorylised with Shrimp Alkaline Phosphatase (SAP) by adding 1 µL SAP to the samples. The ligation was done with an equimolar ratio of plasmid to insert of 1:3 and at presence of 2 µL 10x ligase buffer and 1 µL of T4 ligase enzyme. After incubation at room temperature for 1 h, the cells were transformed in E. coli KRX cells. But the transformation was not successful.
Thursday July 12th
- Team Shuttle vector PCR for more amplification product of 3AOX1 for the part shuttle vector ([http://partsregistry.org/Part:BBa_K863204 K863204]) and 5AOX1 for the part [http://partsregistry.org/Part:BBa_K863200 K863200] was done as described before.
Friday July 13th
- Team Student Academy:
- The groups from Wednesday analyzed their plates. Together with the pupils we made a final analysis of the results, discussed about all problems and questions and helped with the preparation of a presentation, they had to hold in front of all instructors and participants.
- Team Shuttle Vector> Phusion PCR of the fragment tAOX1 that contains upstream an bidirectional double AarI restriction site was done with the wild type X-33 and GS115 as DNA template. The positive control was as ever positive, but no other amplicons.
- Team Cloning of Bacterial Laccases:
- We got new Streptomyces DNA from or supervisor C. Rueckert. He did an alignment with our S. griseus and S. lavendulae sequences against a local database for Streptomyces and identified S. rosechromogenes, S. tuebingen and S.goettingen. Those laccase genes showed simularity to our laccase genes. Since he had isolated chromosomal DNA we were able to work with them. We set the PCR with the three Streptomyces. On this PCR S. tuebingen DNA could be amplified, but it wasn't specific for that what we expected. No products could be amplified on the other two.
Saturday July 14th
Sunday July 15th
Summary of Week 12
hier eine Zusammenfassung
Week 12 (07/16 - 07/22/12)
Contents |
Weekly Seminar
- For our sponsoring we sent all the treaties, to all our sponsors so far.
- Julia V. will organize a guided tour to a sewage treatment plant.
- The summer school was a great success and all the pupils appreciated our organization and the experiments.
- Julia V. will be responsible for the wiki design from now on.
- A big question appeared: How can we put our database on the wiki?
- We will take new team photos next week.
- For the safety/security everyone has to read the safety regulations regarding the organisms and chemicals.
- Next week there will be a short presentation about all used organisms and their laccases
- Agatha and Saskia will create templates for our webpages.
- Nadine will gather information about places to stay in Amsterdam and how to get there.
- Everyone has a passport?
- Gabi will present the judging criteria next week.
- Julia is looking for a BioBrick, which we could use and improve.
- Isabel created a poster for the conference in Munich, everyone has to review it.
Monday July 16th
- Team Fungal and Plant Laccases:
- After we forgot to delete the signal peptide sequences, which are present in the fungal laccases we designed new forward primers for the laccases tvel5, tvel10, tvel13, tvel20 and pcil35 with the overhanging ends for cloning in our shuttle vector. Trametes laccases have a signal sequence after the start codon. This signal peptide we now delete by taking the first 20 bases after this sequence in our FW primers.
- Our plants had a great time during the last weeks in the climate chamber. So today it was time for them to donate their seeds for RNA isolation, cDNA synthesis and a PCR (check protocols). We ran an additional sample with actin primers as a positive control. However both samples did not show any bands. Maybe the high salt concentration in our sample was responsible or the laccase concentration in the 1:10 diluted cDNA was too low. We will do some washing and try again.
- Team Cellulose Binding Domain:
- The two plasmids (p570 and p714) with the cellulose binding domains of [http://www.ncbi.nlm.nih.gov/nucleotide/327179207?report=genbank&log$=nucltop&blast_rank=3&RID=152ZCN0E01N Cellulomonas fimi ATCC 484 exoglucanase gene] and the [http://www.ncbi.nlm.nih.gov/nuccore/M73817 cellulose binding protein gene (cbp A)] from the fermentation group of Bielefeld University arrived.
- Team Cultivation & Purification:
- We searched for some information for the best cultivation conditions in the internet. We found an interesting report of the [http://www.dbu.de/OPAC/ab/DBU-Abschlussbericht-AZ-13191.pdf Deutsche Bundesstiftung Umwelt (DBU)] containing some interesting facts about different laccases as for example that the bacterial laccases are toxic to the bacterias, so that the production could be better under oxygen limitation and reduced temperature. Based on this article we decided to test flask with and without baffles and different temperatures.
- We prepared the basic media and solutions we will need in the lab.
- Note: All following BioBricks are cloned into pSB1C3 and therefore cultivated with 20 µg/mL chloramphenicol unless otherwise specified! Cultivations of E. coli KRX without plasmid will be performed without antibiotics.
Tuesday July 17th
- Team Site Directed Mutagenesis: Made Clone Manager files for the Trametes versicolor laccase plasmids we got send and analyzed them:
- tvel5:
- one silent mutation (at 154 bp) and one amino acid alternating mutation at 227 bp (G for A, replacing D with N), a third was claimed to be at 1559 but sequencing showed it wasn’t.
- No illegal restriction-site for the Silver-assembly, but three Freiburg-assembly restriction-sites: two NgoMIV- and one AgeI-restriction-sites
- tvel10:
- eight silent mutations 171, 444, 1020, 1173, 1239, 1443, 1485 & 1503 bp
- Two amino acid alternating mutations (1048 bp C for T, replacing F with L and 1078 bp A for G, replacing D with N)
- Two PstI-restriction-sites (One in the signaling-sequence at 7 bp and one at 1160) and one SpeI-restriction-site at 241 bp
- One Freiburg-assembly restriction-site (AgeI at 912 bp)
- tvel13:
- 56 silent mutations
- Three amino acid alternating mutations and one whole codon is missing at the very end
- One EcoRI- and one PstI-restriction-site
- Two Freiburg-assembly restriction-sites (two NgoMIV-sites)
- tvel20:
- about 32 silent mutations, three amino acid alternating mutations and four Freiburg-assembly restriction-sites (one AgeI, three NgoMIV)
- pcil35:
- no illegal restriction-site
- tvel5:
- Team Cultivation & Purification:
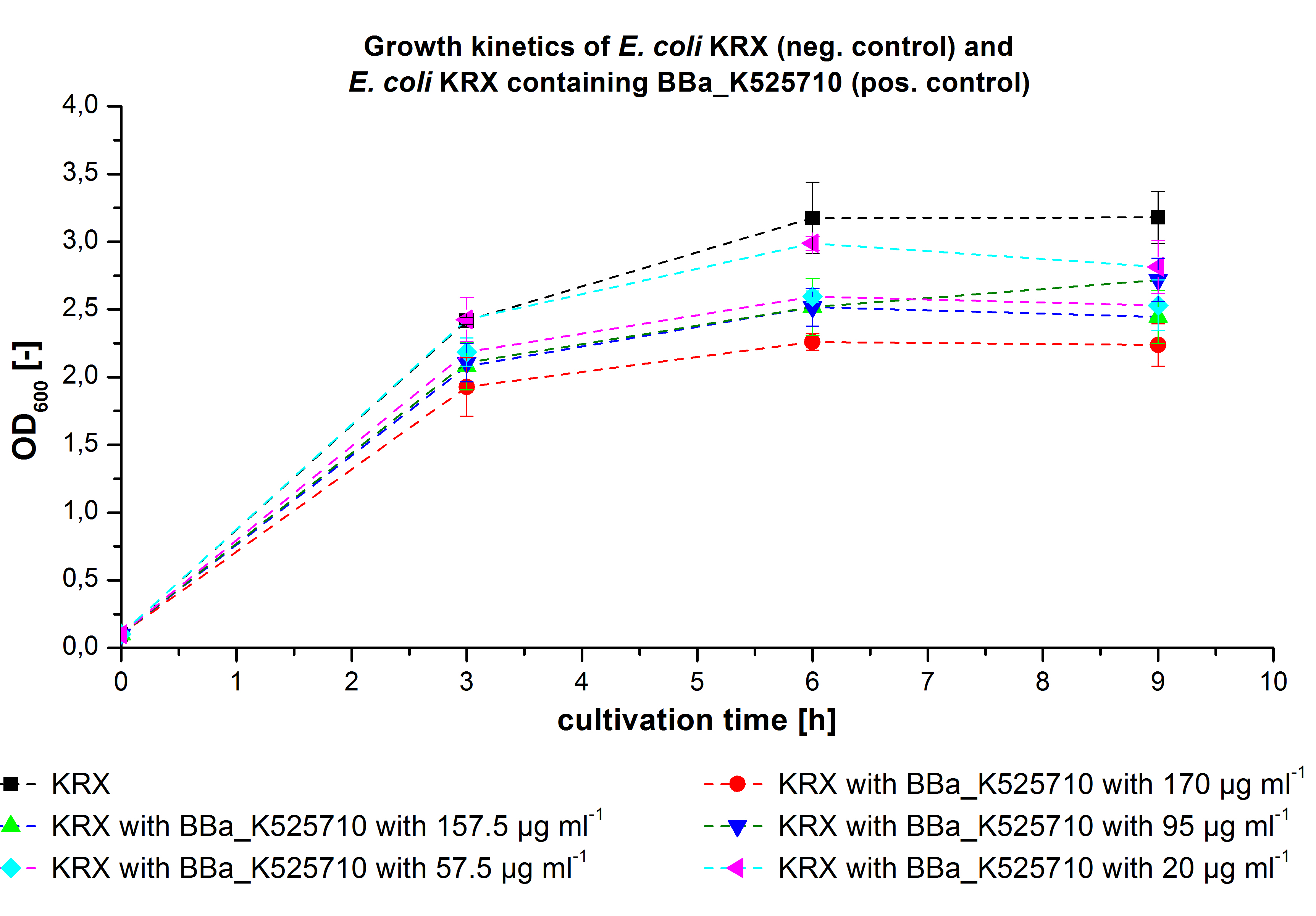
- We prepared our first precultures of E. coli KRX as negative control and E. coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and pBpL6.
- Note: We will cultivate E. coli KRX with pBpL6 until we will get the laccase ORF with T7 promotor and His tag in the same pSB1C3 vector as the other BioBricks. To cultivations of E. coli KRX with pBpL6 we always will add 100 µg/mL ampicillin.
Wednesday July 18th
- Team Site Directed Mutagenesis: Made Primer-Mixes for the bacterial laccases. Set Pre-culture of XL1 blue. Got everything ready for Lab work.
- Team Cloning of Bacterial Laccases:
- We diluted our chromosomal DNA to a concentration of 20 ng/µL, since the volume we got was to low for doing many PCRs and did again a PCR reaction. This time S. goettingen laccase DNA could be identified but not S. tuebingen. The reaction conditions were the same so we were surprised because S. tuebingen didn`t work.
- Team Fungal and Plant Laccases: Some of the ordered parts from the Parts Registry arrived and we plated the biobricks [http://partsregistry.org/Part:BBa_K500000 BBa_K500000], [http://partsregistry.org/Part:BBa_K500001 BBa_K500001], [http://partsregistry.org/Part:BBa_K500002 BBa_K500002], [http://partsregistry.org/Part:BBa_K500003 BBa_K500003] and [http://partsregistry.org/Part:BBa_K392014 BBa_K392014]. Above all we are interested in [http://partsregistry.org/Part:BBa_K500002 BBa_K500002] because it’s a codon optimized laccase from Trametes versicolor and we want to use this laccase in our P. pastoris shuttle vector and characterize it.
- Team Cultivation & Purification:
- Today we performed our first flask cultivation. We cultivated E. coli KRX with [http://partsregistry.org/Part:BBa_K500005 BBa_K500005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], pBpL6 and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and as negative control we used E. coli KRX.
- Settings: We used 300 mL flasks without baffles, final volume: 60 mL, autoinduction medium, 30/37 °C, durance: 24 hours
- Found out that we had a mixed culture of E. coli KRX with pBpL6, because it grew on chloramphenicol, but has only an ampicilline resistance. So we could not use this culture.
- Today we performed our first flask cultivation. We cultivated E. coli KRX with [http://partsregistry.org/Part:BBa_K500005 BBa_K500005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], pBpL6 and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and as negative control we used E. coli KRX.
Thursday July 19th
- Team Site Directed Mutagenesis:
- Made electrocompetent XL1 blue cells. Made pfu-PCR of the bpul-plasmid with bpul-a2883t primer-mix, the ecol-plasmid with ecol-g2307a primer-mix, the xccl-plasmid with the xccl-g2247c primer-mix and tthl-plasmid with the tthl-g2796a primer-mix. Digested the template with DpnI.
- Generated Primers for silent mutations of tvel10 illegal restriction-sites:
- the illegal SpeI will be deleted by changing acT to ggA (Threonine) at 243 bp of the know sequence
- the illegal PstI will be deleted by changing ccT to ccA (Proline) at 1161 bp of the known sequence
- one illegal restriction site in the signaling-sequence can not be mutated, since it is to close to the beginning of the known sequence (7 bp) and also it is not essencial, because the gene will be used without the signaling sequence
- Team Cellulose Binding Domain:
- Gathered some information about carbohydrate binding domain X2 (which is a more common domain in the organisms we handle than a cellulose Binding Domain).
- Used NCBI Nucleotid BLAST on <partinfo>BBa_K392014</partinfo> (the Cellulose-binding motif from C. josui Xyn10A gene) and it 100% fits to 1526 bp to 2608bp of AB041993.1 (Clostridium josui xynA gene for xylanase A).
- Used NCBI Protein-BLAST on AB041993.1 and found one cellulose binding domain and one carbohydrate binding domain within the protein.:
- pfam02018: CBM_4_9 (Carbohydrate binding domain) from AS 193 to AS 332 - corresponds to 1088 bp - 1507 bp in the gene and is not in the coding sequence of <partinfo>BBa_K392014</partinfo>.
- cd09619: CBM9_like_4 (DOMON-like type 9 carbohydrate binding module) from AS 716 to AS 887 - corresponds to 2657 bp to 3172 bp in the gene and is also not in coding sequence of <partinfo>K392014</partinfo>.
- Given sequence of <partinfo>BBa_K392014</partinfo> (1589bp to 2590bp) does corresponds to the sequence of the Glycosyl hydrolase 1589 bp to 2590 bp and is not one of the binding motifs. The whole review can be read at the [http://partsregistry.org/Part:BBa_K392014:Experience Experience-page of BBa_K392014].
- Team Cloning of Bacterial Laccases:
- S. goettingen and S. tuebingen DNA were cleaned up and we done an enzymatic digestion to ligate it into the pSB1C3 vector. We did the digestion with EcoRI and SpeI.
- Team Fungal and Plant Laccases: Colonies from plated BioBricks from 18.07 were spread on nutrient agar for plasmid isolation.
- Team Cultivation & Purification:
- The cultures of 07/18 were centrifugated, cells were disrupted via sonication in the special buffer for each laccase and after another centrifugation the supernatant was given to the activity test team.
- Made precultures analogous to them on 07/17.
Friday July 20th
- Team Modeling: We need a contact to a clarification plant to get information about clarification plant itself and perhaps to proof our cleaner with real probes. therefor we are calling Mr. Bülter form the clarification plant Schloß Holte (near Bielefeld) and he invited us to present our project.
- Team Shuttle vector: Gel purification of the fragments Kozak-MFalpha1 [http://partsregistry.org/Part:BBa_K863206 K863206] and 3'AOX1 [http://partsregistry.org/Part:BBa_K863201 K863201] was done.
- Team Site Directed Mutagenesis:
- Transformation of XL1 Blue with the PCR products (see day before)
- Team Cellulose Binding Domain:
- Used Protein-BLAST on the translated sequence of the Clostridium cellulovorans cellulose binding protein, the Bacillus halodurans strain Cochin chitinase GU481106.1 and chitin-binding protein [Bacillus halodurans C-125] BAB05022.1, to get more information about the predicted sequences of Carbohydrate binding domains.
- Looked up possible linkers in the Parts Registry:
- 2 aa GS linker: [http://partsregistry.org/Part:BBa_J18920 BBa_J18920] [GS]
- 10 aa [GS]x linker: <partinfo>BBa_J18922</partinfo> [GSGSGSGSGS]
- 15 aa flexible glycine-serine protein domain linker; Freiburg standard 1 Star: <partinfo>BBa_K157013</partinfo> [GGGGSGGGGSGGGGS]
- 10 aa flexible protein domain linker 1 Star: <partinfo>BBa_K105012</partinfo>: [GENLYFQSGG]
- Made a few possible primers for [http://partsregistry.org/Part:BBa_K863112 CBDclos] with T7 and RBS (<partinfo>BBa_K525998</partinfo>), a Freiburg-Suffix and possible linkers
- Wrote an E-Mail to Jun.Prof Thorsten Seidel (FRET-Expert of Bielefeld University) asking him about the linkers they use for fusing GFP for FRET.
- Team Fungal and Plant Laccases: Isolating the plasmids [http://partsregistry.org/Part:BBa_K500000 BBa_K500000], [http://partsregistry.org/Part:BBa_K500001 BBa_K500001], [http://partsregistry.org/Part:BBa_K500002 BBa_K500002], [http://partsregistry.org/Part:BBa_K500003 BBa_K500003] and [http://partsregistry.org/Part:BBa_K392014 BBa_K392014]. To be sure that in every plasmid contains the correct part we made a control restriction with NotI. It showed that all parts are on the correct hight in agarose gel.
- Team Cultivation & Purification:
- Another flask cultivation was made analogous to the one on 07/18 but at 23 °C.
Saturday July 21st
- Team Cellulose Binding Domain:
- Made primers for [http://partsregistry.org/Part:BBa_K863121 GFP_His] (using [http://partsregistry.org/Part:BBa_I13522 BBa_I13522] as template)adding a C-termial His6tag for easy extraction
- Jun.Prof Thorsten Seidel answered to the mail of yesterday and said that GFPs are easy to fuse and that they use linkers with four AS:
- ccg gtc gcc acc upstream of the GFP
- gaa agc ggc cgc downstream of the GFP; Which is will work fine, even with a proline in it's sequence.
- We decided to use four linking AS also and chose to add a C-terminal Glycine-Serine-Linker ([http://partsregistry.org/Part:BBa_J18920 BBa_J18920]) to the cellulose binding domain, which would be four linking AS with the Freiburg-scar (Threonine-Glycine).
- Team Cloning of Bacterial Laccases:
- Since our PCRs didn't work and the Streptomyces laccases have more then 70% GC content we changed the dNTP concentration to 10 mM per base. Our gelelectrophoresis showed us that again S. goettingen has a product in correct length so we did again a PCR with same conditions. The rest of this product was cutted from an agarose gel and set ligation with the pSB1C3 vector and transformation. Because the isolated DNA concentration was to low the transformation showed no positive results.
- Team Cultivation & Purification:
- Culture from 07/20 was harvested and the cells were disrupted via sonication.
Sunday July 22nd
- Team Shuttle Vector:
- To get more amplicon from the part 5AOX1 [http://partsregistry.org/Part:BBa_K863200 K863200] an other Phusion PCR was done and the PCR product gel purified.
- To get the tAOX1 fragment for the shuttle vector, we run a PCR with the KOD DNA polymerase as descriped in the protocol.
- While debugging and finding an solution for PCR fragment for the part tAOX1, test the annealing temperature of 60 °C that was proposed by the [http://www.neb.com/nebecomm/tech_reference/tmcalc/default.asp#.UGNW-K-zZ5w Tm calculator from NEB]. Therefore, we run a temperature gradient from 50 °C until 67 °C with the usual protocol for Phusion DNA polymerase. But no specific product could be detected.
- Because there were also many problems to get a PCR fragment of the HIS4 fragment for the shuttle vector we run a temperature gradient from 55 °C until 60 °C with the usual Phusion DNA polymerase protocol. From 50 °C until 65 °C we could detect the right fragment. The highest yield was at 60 °C.
- Team Cellulose Binding Domain:
- We decided to use the exact sequence of [http://partsregistry.org/Part:BBa_J18920 BBa_J18920] (GGCAGC) of the linker in the partsregistry.
- Checked the CBDs we had access to for illegal restriction sites and found none, not even NgoMIV or AgeI. So we decided to use a Freiburg-Assembly to build the CBD-GFP fusion protein.
Summary of Week 13
hier eine Zusammenfassung
Week 13 (07/23 - 07/29/12)
Contents |
- From 07/23 - 07/25/12, some of our team members participated at the CAS conference in Munich.
Weekly Seminar
- we will attend at the Biolympics it Münster, hosted by the [http://bts-ev.de/ bts] just to have some fun.
- We decided to write down every result in the labjournal, so that problems and their solution can be understood and reconstructed
Monday July 23rd
- Team Activity Tests: Today was the moment of truth. We received the first recombinant laccases from the cultivation team. We started with testing the supernatant from the cultivated cells to check whether they secrete laccase. We added 0,1 mM ABTS in each well that was filled with supernatant from E.coli KRX that contained the plasmid to produce ECOL, BPUL, BHAL, XCCL or E. coli KRX (negative control) and measured as usual. There was no change in the OD so that we assume that no secretion takes place.
- Team Shuttle vector:
- Electroporation of RFP::pSB1C3 was done to get more plasmid DNA, but the wrong competent cells and selective plates were choosen.
- Phusion PCR of the tAOX1 part for the shuttle vector was done without betaine addition with the genomic DNA of P. pastoris GS115 and the wild type X-33. But only a small unspecific product was visible.
- Team Cloning of Bacterial Laccases: The transformation from the 21st July showed no results. The plates were unfortunately empty. So we did an another PCR. This time the Annealing Temp. was set to 56°C.
- Team Fungal and Plant Laccases: Successful PCRs of laccase gene Tv5 from Trametes versicolor with plasmid DNA from Uni Greifswald as template.
- Team Site Directed Mutagenesis: Transfered 4 colonies of each transformation to an new dish for plasmid-isolation.
Tuesday July 24th
- Team Shuttle Vector: To get the fragment tAOX1 for the shuttle vector the Phusion PCR conditions were changed. A two-step PCR was done with the following conditions: After initial denaturation at 98 °C for 6 min, the first 10 cycles were run at: denaturation at 98 °C for 15 s, primer annealing at 55 °C for 30 s, elongation at 72 °C for 30 s. The following cycles were run at: denaturation at 98 °C for 15 s, primer annealing at 67 °C for 30 s, elongation at 72 °C for 30 s. The final elongation run at 72 °C for 5 min. But the PCR ghost was not satisfied and the right amplicons could not be detected. Just some unspecific small fragments could be detected in the geleletrophoresis.
- Team Site Directed Mutagenesis: Plasmid-isolation of all 16 different colony-dishes
- Team Fungal and Plant Laccase: Since our cDNA is now ready to go and also our primers already arrived we started a PCR. Check protocols for more information about the exact setup. Unfortunately no bands could be seen via gel electrophoresis. We blamed the high salt concentration and decided that a some cleaning via ethanol precipitation might be a solution to our problem.
- Team Activity Tests: After testing the supernatant of the cultivations yesterday we got to the more exciting part today. We tested the cell extract for laccase activity by filling each well with 140 µL cell extract, 40 µL buffer, 18 µL H2O and added 2 µL ABTS. Unfortunately no activity was seen. We felt like we could not take the laziness of our laccases. We thought and discussed what the reason could be for them to be inactive.
- Is there something wrong with the transformed construct, maybe a mutation?
- Was the laccase synthesized but is inactive for some reason?
- Was the laccase produced but not in a high enough amount so that we can´t detect its activity in the first place?
Another thought was that there might be copper missing. Since laccase is a copper-enzyme it might be necessary to add copper to the medium whenever it is produced in such high amounts. It´s late, so we will do that tomorrow.
Wednesday July 25th
- Team Site Directed Mutagenesis: Test-digestion (with the enzym of the mutated restriction-site) of the Plasmids from the 16 different colonies revealed that two bpul, one ecol and two tthl mutants have the right bands in the gel and seem be mutated correctly. Xccl-colonies are all unmutated (no Band at 3636).
- pfu-PCRs with the two correct bpul-plasmids as template and the bpul-g2317t primer-mix
- two new pfu-PCRs with the xccl-plasmid one with the g2247c primer-mix and the other with the xccl-g3633c primer-mix
- Prepared all positive colones for sequencing
- Team Cultivation & Purification:
- We made precultures of E. coli KRX without plasmid and with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] , pBpL6 as well as with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020] or [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015].
- Team Activity Test: Today is copper day! First we divided each sample (the ones from yesterday) and added 2mM or 4mM copper sulfate to the halfs, respectively. We incubated our samples for 2h and hoped that the concentration gradient would cause the laccases to exchange the ion they have integrated instead against copper. We measured their activity again after those 2h but there was no change seen. Team Cultivation plans to add copper from the very beginning of cultivation next time.
Thursday July 26th
- Team Modeling and Team Sponsoring: meeting Mr. ??? from the clarification plant of Schloß Holte and finding a new cooperation partner. He want to give us the information we need to equate our model and design our cleaner. On top he wants to ask if the clarification plant can sponsor our project.
- Team Site Directed Mutagenesis: Went on with the PCR products of xccl and bpul, transformed them into XL1 Blue and plated them on select-agar
- pfu-PCRs with the tvel10-plasmid, one with the tvel10-243 primer-mix and one with the tvel10-1161 primer-mix. Both PCRs showed correct bands for the PCR-product in a test-agarose-gel-electrophoresis. Digested template with DpnI.
- Team Fungal and Plant Laccases: We did PCR on Trametes versicolor laccase tvel5 and Pycnoporus cinnabarinus pcil35 with Tv_lac10.P.FW / Tv_lac10.S.RV and Pc_lac35.P.FW / Pc_lac35.S.RV primers. Additionally we want the laccases with overhanging ends for cloning in our shuttle vector for expression in Pichia pastoris. Therefore we used Pc_lac35_FW_oS / Pc_lac35_RV and Tv_lac5_FW_oS / Tv_lac5_RV primer pairs.
- Team Cultivation & Purification:
- Glycerine cultures were made of all given BioBricks, to use them for precultures.
- Made new precultures analogous to 07/25, as well as a preculture of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010].
- Found out, that it could be important to add CuCl2 to the medium, because the copper is important for the active center of the laccases and to improve their stability.
Friday July 27th
- Team Site Directed Mutagenesis: plated four colonies of each transformation-dish (xccl & bpul) on a petri dish for plasmid-isolation. Purified the digested PCR-products of tvel10.
- Team Cellulose Binding Domain:
- We made the decision to only use cellolose binding domains (CBDs) and not carbohydrate binding domains of any kind to keep them comparable. This means we won't use the binding domains of Bacillus halodurans.
- Redesigned Primers for [http://partsregistry.org/Part:BBa_K863112 CBDclos] - added the bases TA in between RBS and ATG since the RBS can be made stronger by adding more adenines in the sequence upstream of the RBS.
- Made similar primers for [http://partsregistry.org/Part:BBa_K863102 CBDcex] (the CBD of the Cellulomonas fimi exoglucanase we got for the fermentation-group of Bielefeld University)
- Checked the Wiki of the iGEM 2012 Osaka University-Team about their <partinfo>BBa_K392014</partinfo> BioBrick which should be a binding motif, but is the glycosyl hydrolase domain, for their intentions. We guess they really messed it up. It would have been nice to have a CBD from the partsregistry.
- Team Fungal and Plant Laccases: The PCR on tvel5 laccase was positive after the first trial for both primer pairs. For the tvel35 laccase the PCR didn’t work.
- Team Cultivation & Purification:
- We cultivated E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015],the new BioBrick [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010] as well as pBpL6. As negative control we used E. coli KRX.
- Settings: 300 mL/ 1 L flasks without/without baffles, final volume: 60 mL/200 mL, autoinduction medium, 0.25 mM CuCl2, 30 °C
- We made glycerine cultures of homologous culture of E. coli KRX with pBpL6.
- We cultivated E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015],the new BioBrick [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010] as well as pBpL6. As negative control we used E. coli KRX.
Saturday July 28th

- Team Shuttle Vector:
- Because the first Gibson assembly failed, more fragments with a higher concentration are necessary. All parts were amplificate by the same protocols as described.
- Also the Part HIS4 [http://partsregistry.org/Part:BBa_K863202 K863202] was amplified as described.
- And the Part tAOX1 [http://partsregistry.org/Part:BBa_K863203 K863203] was amplified by the same conditions as the fragment tAOX1 for the shuttle vector.
- Team Activity Tests: See, we are back already! Today we received different samples from [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 ECOL], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BPUL], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863022 BHAL], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 TTHL], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 XCCL] and E. coli KRX (negative control - cells without plasmid). The cells were all cultivated at 30°C and supplied with copper. We tested each one of them but none of them showed laccase activity.
- Team Fungal and Plant Laccases:
- Purification of Trametes versicolor tvel5 PCR product.
- With the cDNA samples we got out of the siliques of A. thaliana we did two PCRs. After waiting a hour for the PCR to stop, we started a gel, hoping for really thick bands. But nonetheless there were non. Also the PCR with actin primers didn't give a result. A possible reason for this could have been contaminations like salts in our samples. Before translating the mRNA into cDNA we measured the following properties: 260/280: 1.93 and 260/230: 1.28. This indicates a high salt concentration which could be interfering during the PCR. For the next PCR we want to wash our cDNA and hope for better results with a clean and pretty sample.
- Team Cultivation & Purification:
- Cells of cultivation 07/27 were disrupted via sonication and given to the activity test team.
Sunday July 29th
- Team Cultivation & Purification:
- Preparation of all the stuff needed for SDS-Pages.
- Made new precultures of E. coli KRX without plasmid and with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015],[http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], as well as pBpL6 and our new BioBrick [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005].
Summary of Week 14
hier eine Zusammenfassung
Week 14 (07/30 - 08/05/12)
Contents |
Weekly Seminar
- From now on, there will be two weekly meetings: monday, 12.15 and thursday 9.00.
- Nadine sent all of us a poll, answers will be used on the wiki.
- For picture uploads we will use the name: Bielefeld2012_x.
- Which sewage treatment plant uses activated carbon in the third stage of treatment?
- On 25th of august, all german teams will host the SynBioDay. In Bielefeld we will cooperate with [http://www.bielefeld-marketing.de/de/index.html Bielefeld Marketing] and to raise public interest we will call it StreetScience. As part of our collaboration we will invite the iGEM team from SDU-Denmark to help us with the organization and the realization of this common day.
- On StreetScience we will have a little experiment about extracting fruit DNA with an easy method.
- Do we want to travel to Amsterdam by bus or train?
- Do we want a ship as our accommodation in Amsterdam?
- Gabi has presented the judging criteria including the bronze, silver and gold criteria as well as the special prices.
Monday July 30th
- Cloning of Bacterial Laccases:
- The PCR products from the 23th July was digested with EcoRI and SpeI same as the pSB1C3 vector. Then the ligation was set and transformed into competent E. coli KRX cells.
- Since we haven't got any more PCR products we set a PCR. Only S. goettingen worked.
- Team Fungal and Plant Laccases: Digestion of tvel5 PCR product with prefix and suffix ends for cloning in pSB1C3 and digest of laccase with the overhangs for cloning in shuttle vector. Additionally we did the PCR on tvel35 with both primer pairs again. This time we lowered the annealing temperatures and got products with both primer pairs.
- Team Cellulose Binding Domain:
- Redesigned the primer-sequences another time giving the CBDs a few additional amino acids from the by Protein-BLAST predicted domain. [http://partsregistry.org/Part:BBa_K863102 CBDcex]: 4 AS N-terminal, 2 C-terminal. [http://partsregistry.org/Part:BBa_K863112 CBDclos]: 2 N-terminal (starting with a natural ATG) and 2 AS C-terminal.
- Prepared <partinfo>BBa_K392014</partinfo> for Sequencing
- Team Site Directed Mutagenesis: Plasmid-isolations of all the bpul- and xccl-mutants.
- Team Cultivation & Purification:
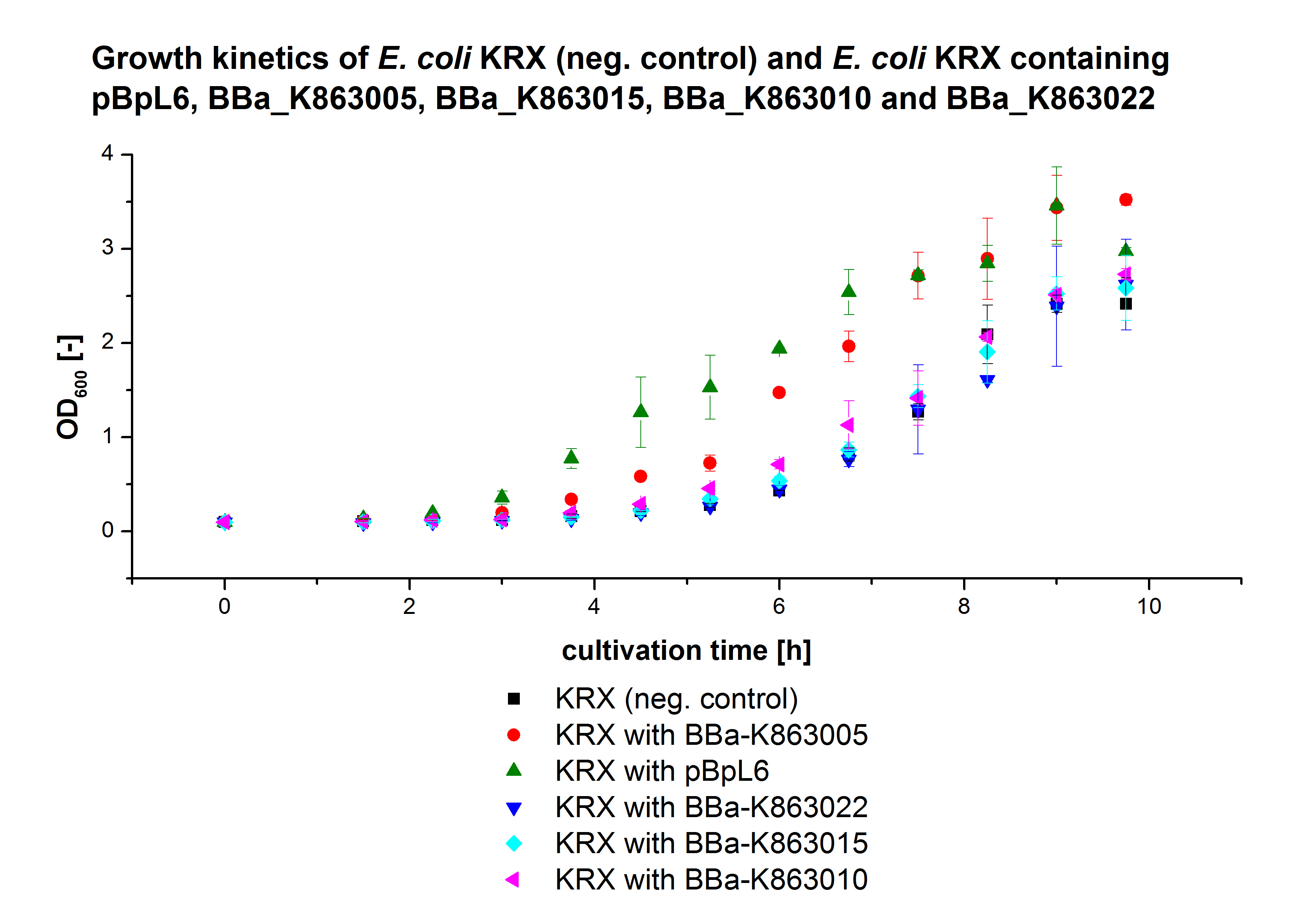 Figure 1: Growth kinetics of cultivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and pBpL6. Parameters: autoinduction medium, 0.25 mM CuCl2, 28 °C.
Figure 1: Growth kinetics of cultivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and pBpL6. Parameters: autoinduction medium, 0.25 mM CuCl2, 28 °C.- We made the SDS-Pages for the cultivation from 07/27, but they did not seem to be promising.
- We started another cultivation of E.coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and pBpL6.
- Settings: 300 mL flasks without baffles, final volume: 60 mL, autoinduction medium, 0.25 mM CuCl2, 28 °C.
- A growth kinetics was recorded every 45 minutes.
- We decided that we also need a positive control for the next cultivations, to see if our autoinduction medium works. We chose [http://partsregistry.org/Part:BBa_K525710 BBa_K525710]. Furthermore we will make cultivations in which we manually induce the expression after 4 hours.
- Made a preculture of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and pBpL6 as well as with [http://partsregistry.org/Part:BBa_K525710 BBa_K525710]. We used E. coli KRX as negative control.
Tuesday July 31st
- Team Shuttle Vector
- To get the tAOX1 fragment for the shuttle vector, we run a primer gradient from 0.1 µM to 1 µM in 0.1 µM steps with a primer annealing time of 15 s and 20 s and the usual Phusion PCR protocol. No fragments could be detected.
- A DNA template gradient from 10 ng to 250 ng in 50 ng steps was choosen. But no fragment could be detected. Also the primer annealing temperature was varied in 15 s and 20 s. But only small unspecific fragments with from 100 bp to 150 bp could be detected.
- Team Cloning of Bacterial Laccases:
- After our laccases are not produced or not detectable we decided to try to express the laccases with a constitutive promoter. Therefore we searched for appropropriate promoters and found different promoters from the [http://partsregistry.org/Ribosome_Binding_Sites/Prokaryotic/Constitutive/Anderson Anderson promoter family]. We picked three different promoters with different promoter strengths. We chose the parts [http://partsregistry.org/wiki/index.php/Part:BBa_J23103 BBa_J23103], [http://partsregistry.org/wiki/index.php/Part:BBa_J23103 BBa_J23110] and [http://partsregistry.org/wiki/index.php/Part:BBa_J23103 BBa_J23117] and for all three promoters the RBS [http://partsregistry.org/wiki/index.php/Part:BBa_J23103 BBa_B0034]. The primers were designed that the FW and the RV primers anneal together to a short oligonucleotide with overhanging restriction site ends for EcoRI and SpeI. So we don’t have to cut the annealed primers, because the sites should appear with correct annealing of FW and RV primers. The goal is to clone the different promoters, the PCR products with the different laccase genes with His-Tag in pSB1C3 in one ligation step. Furthermore we want to exclude the possibility that the His-tag is the reason for no activity, so we also want to clone the gene sequences without a His-tag under control of a constitutive promoter in pSB1C3.
- Additionally we want to produce the constructs with a new T7 promoter. After our laccases were not expressed we now think that maybe the RBS <partinfo>BBa_B0034</partinfo>, which we changed in our primers from originally 5' aAagaggagaaa 3' to 5' aGagaggagaaa 3' is not or poorly recognized from the ribosomes in the cells.
- We have a primer pair from last year with the T7 promoter sequence with the RBS BBa_B0034. The primers can be annealed to an oligonucleotide. After boiling the primers and cooling down there should be an oligonucleotide with an EcoRI and a PstI restriction site. We now want to assemble the promoter the laccase genes and the pSB1C3 vector in one step. The pSB1C3 backbone was digested with EcoRI and PstI, the laccases with XbaI and PstI, and the promoter has the restriction sites SpeI and EcoRI (we have to digest the promoter with SpeI, because the original restriction site is PstI).
- Team Fungal and Plant Laccases:
- Purification of tvel35 PCR product and digestion for cloning in pSB1C3 backbone.
- Ligation of tvel5 laccase with pSB1C3 backbone.
- Team Site Directed Mutagenesis: Plated four colonies per dish of tvel-t243g and tvel-t1161a for plasmid-isolation.
- Test-restriction of the xccl-mutants showed that no colony was mutated correctly
- The second site directed mutagenesis of bpul-g2317t could not be rated by test-digestion, since the mutation at 2317 is not because of a illegal restriction site, but a amino acid alternating mutation.
- Made four bpul-colonies (with both mutations) ready for sequencing
- plated a four more colonies of xccl (both SDM-sites) for plasmid-isolation
- Team Cultivation & Purification:
- Made SDS-Pages of cultivation from 07/30.
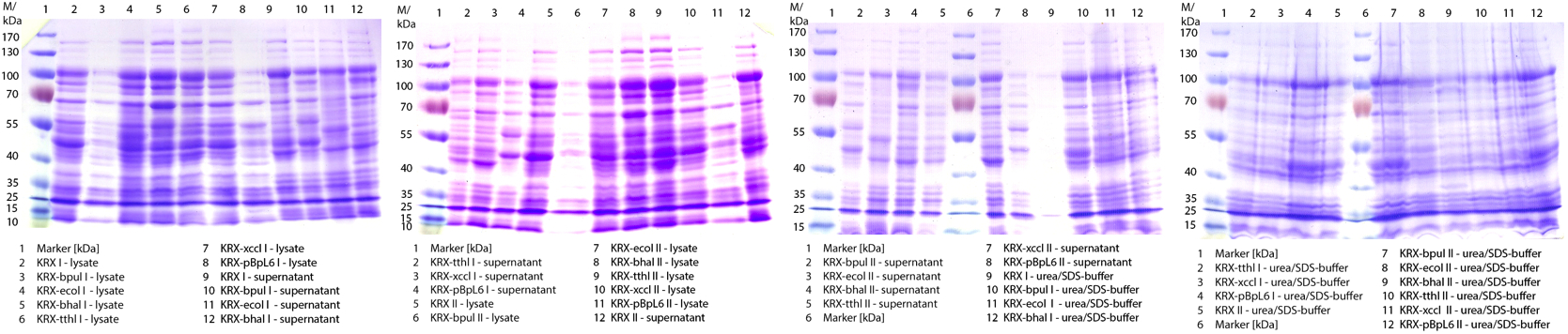 Figure 2: SDS-PAGES of the lysate, the supernatant, and with urea treated pellet (cultivation 07/30) of E.coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] or pBpL6. 60 mL in 300 mL flasks without baffles with autoinduction medium, 0.25 mM CuCl2, 28 °C
Figure 2: SDS-PAGES of the lysate, the supernatant, and with urea treated pellet (cultivation 07/30) of E.coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] or pBpL6. 60 mL in 300 mL flasks without baffles with autoinduction medium, 0.25 mM CuCl2, 28 °C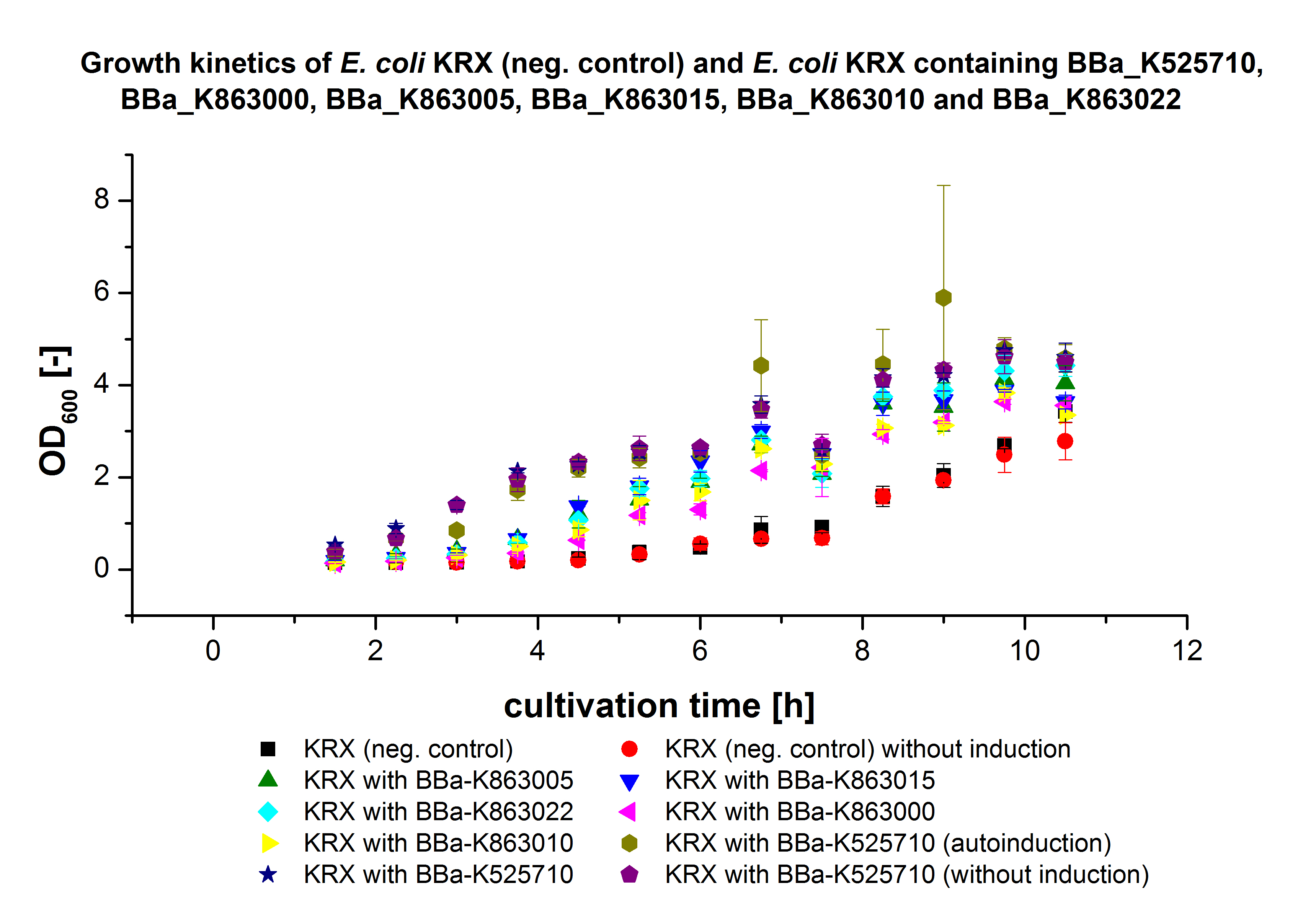 Figure 3: Growth kinetics of cultivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and pBpL6. Parameters: autoinduction or manual induction, 0.25 mM CuCl2, 30 °C.
Figure 3: Growth kinetics of cultivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and pBpL6. Parameters: autoinduction or manual induction, 0.25 mM CuCl2, 30 °C. - Cultivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and pBpL6 as well as with [http://partsregistry.org/Part:BBa_K525710 BBa_K525710]. We used E. coli KRX as negative control.
- general settings: 300 mL flasks without baffles, final volume: 60 mL, 30 °C
- special settings:
- Made SDS-Pages of cultivation from 07/30.
- a) E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/Part:BBa_K525710 BBa_K525710], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] and pBpL6 were cultivated in LB media and manually induced after 4 hours with 0.1 % rhamnose.
- b) E. coli KRX without plasmid was cultivated with LB medium and induced after 4 hours with 0.1 % rhamnose.
- c) E. coli KRX with [http://partsregistry.org/Part:BBa_K525710 BBa_K525710] was cultivated equally to b)
- d) E. coli KRX without plasmid was cultivated with autoinduction medium and 20 µg/mL chloramphenicol as well as 100 µg/mL ampicillin as a control.
Wednesday August 1st
- Team Site Directed Mutagenesis: Plasmid-isolation of tvel- and the xccl-colony-dishes.
- Test-restriction with showed correct bands for two tvel and one xccl-colony.
- Made the correct xccl-plasmid ready for sequencing.
- Team Cloning of Bacterial Laccases:
- With the new T7 promoter we started new assemblies. We first want to try the new promoter with ecol. So wedigested our laccase PCR products from ecol with and without HIS tag for suffix insertion with XbaI and PstI, boiled and cooled down the joined T7 primer pairs and digested pSB1C3 backbone with EcoRI and PstI. If the plan works, the parts anneal to our final plasmid. We did the ligation and transformation of the approach into competent E. coli KRX cells.
- Team Cultivation & Purification:
- Harvesting and cell disruption via sonication of yesterday's cultivation.
Thursday August 2nd
- Team Wiki: This morning we met for taking a new picture of our group and individual pictures of everyone. Check out our beautiful team members here.
- Team Cloning of Bacterial Laccases:
- There were just red colonies on the plates from the transformation the day before. So we didn't do colony PCR and have to do the assembly again.
- Our Laccases genes from S. goettingen, S. tuebingen and S. roseochromogenes are around 2kb. The NEB-Phusion has a range of 1kb/30sec so we thought if we set the elongation time higher we might have products for S. tuebingen,S. roseochromogene so we did PCR. But the results showed that it doesn't depend on the elongation time from the Phusion because we haven't had any products.
- Team Fungal and Plant Laccases: Ligation of tvel5 and pcil35 in pSB1C3.
- Team Site Directed Mutagenesis: pfu-PCRs; two with the positive tvel-t243g-plasmids as templates and the tvel-t1161a primer-mix and another pfu-PCR with the positve xccl-g2247c-plasmid and the xccl-g3633c primer-mix
- Team Cultivation & Purification:
- Made SDS-Pages of cultivation from 07/31.

Friday August 3rd
- Team Cloning of Bacterial Laccases:
- Digest of ecol PCR products with and without HIS-Tag and ligation with pT7 in pSB1C3. Transformation of this ligations and transformation of [http://partsregistry.org/Part:BBa_K525710 BBa_K525710] for Team Cultivation.
- We did an annealing temp. gradient PCR with the Streptomyces bacteria. But expect 'S.goettingen neighter S. tuebingen not S. rosechromogenes did not work.
- Team Fungal and Plant Laccases: Transformation of tvel5 and tvel35 in pSB1C3 backbone.
- Team Site Directed Mutagenesis:
- DpnI-digestion of the pfu-PCR-products (from yesterday). Transformed tevl- and xccl-double-mutants into XL1 Blue and plated them on selection-agar.
- Sequencing results:
- Both tthl-g2796a-mutations were successful
- ecol-g2307a has additional mutations
- Team Cellulose Binding Domain: Transformed the plasmids p714 (with the CDBcex domain) and p570 (with the CBDclos domain) we got from the fermentation group of Bielefeld University in KRX an plated them on kanamycin-selection-agar.
- Team Activity Tests: Our lovely Team Cultivation started new cultivations on 07/30 and we got the products. More precisely we got the supernatant of the lysis Team Cultivation did before. We tested 198 µL of each sample with 2 µL ABTS for the activity reaction, but sadly we couldn't detect anything (see Fig. 5).
Saturday August 4th
- Team Cloning of Bacterial Laccases:
- We did the transformation of the ligations from August 1st again.
- Team Site Directed Mutagenesis: Plated four double-mutant-colonies from each transformation (both tvel-mutants and the xccl-mutant on selection-agar for plasmid isolation.
- Team Cellulose Binding Domain:
- The transformation of p570 brought up only a few colonies, so we took a few and plated them again on a selection-agar-dish.
- Plasmid-isolation of p714
- Team Cultivation & Purification:
- We prepared precultures of E.coli KRX without plasmid and E.coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], pBpL6 or [http://partsregistry.org/Part:BBa_K525710 BBa_K525710] as positive control.
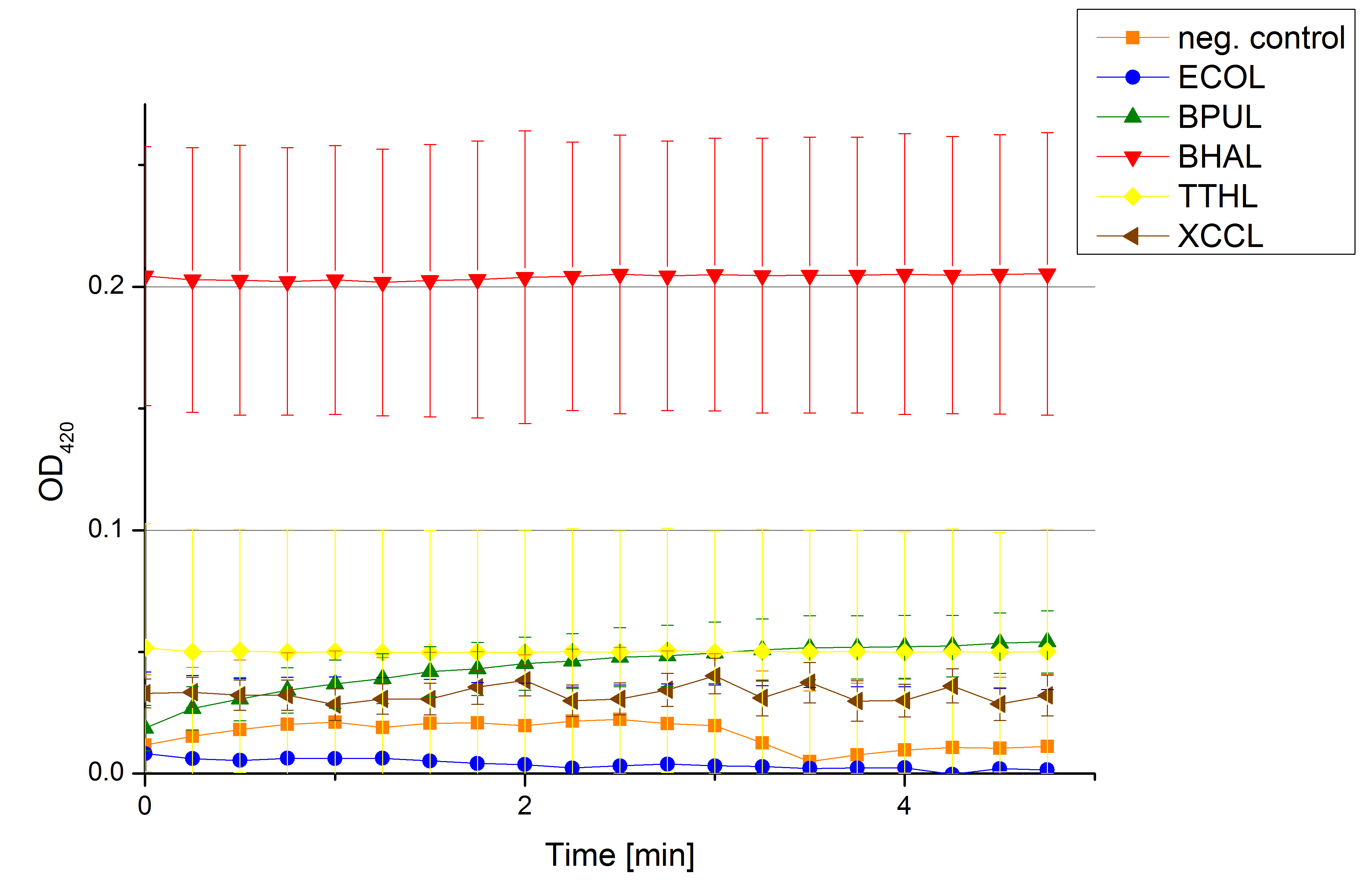
- Team Activity Tests: Another cultivation ended and we are happy to measure the supernatants from the 31st September cultivated samples. We used 158 µL of the supernatant with 40 µL and added 2 µL of 20 mM ABTS to measure some activity (Fig. 6). Nevertheless we couldn't see any increase in absorbency at 420 nm after 5 minutes, indicating, that we have no active laccase in the supernatants.
Sunday August 5th
- Team Cloning of Bacterial Laccases:
- The assemblies don't work by now in one step by now, we always get just red colonies from the original RFP plasmid. So we started to clone one part after another in pSb1C3 backbone. For this reason we want to ligate the ecol gene without any promoter in pSB1C3 backbone and in the next step do a prefix insertion with the promoter fragment. Therefore we did the restriction of ecol and ecol_HIS PCR products, the ligation in pSB1C3 and the transformation.
- Team Site Directed Mutagenesis: Plasmid-isolation of the xccl- and tvel-double-mutants.
- Test-restriction showed two positive plasmids on the xccl-g2247c and xccl-g3633c-mutagensis and no positve tvel.
- plated two colonies of the ecol-g2307a-mutagenesis and plated them on select-agar for plasmid-isolation.
- Prepared the two positive double mutants of the xccl-plasmid for sequencing.
- Team Cellulose Binding Domain:
- Transformated BBa_I13522 in KRX and plating it on AMP-selection-agar.
- Plasmid-isolation of p570
- PCR of [http://partsregistry.org/Part:BBa_K863103 CBDcex] (417 bp) and [http://partsregistry.org/Part:BBa_K863113 CBDclos] (369 bp) followed by test-gel-electrophoresis showed bands of the correct size and PCR-Clean-up
- Restriction of PCR-products and pSB1C3_RFP with XbaI and PstI.
- Team Cultivation & Purification:
- We made another flask cultivation of E. coli KRX without plasmid and with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], pBpL6 and [http://partsregistry.org/Part:BBa_K525710 BBa_K525710] as positive control.
- general settings: 250 mL flasks without baffles, final volume: 50 mL, LB medium
- additional settings:
- We made another flask cultivation of E. coli KRX without plasmid and with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], pBpL6 and [http://partsregistry.org/Part:BBa_K525710 BBa_K525710] as positive control.
- a) 2 flasks of each culture were inducted with 0.1 % rhamnose after 4 hours of cultivation
- b) 2 flasks of each culture were not induced.
- We recorded the growth kinetics once per hour.
Summary of Week 15
hier eine Zusammenfassung
Week 15 (08/06 - 08/12/12)
Contents |
Weekly Seminar
- We did a safety and security training today.
- We agreed to write in wiki code in our wiki, because not everyone is capable of writing in html.
- Next week Hakan will present his problems with getting the PCR to work
Monday August 6th
- Team Cloning of Bacterial Laccases:
- No positive colonies after transformation of our assemblies from August 1st but we realized that the primers we used for making the promoter parts can’t ligate with our backbone because the primers are dephosphorylated and the plasmid backbone is dephosphorylated, too. Much effort in a mission which can't work but at least we know now why it doesn't work.
- Picking positive colonies from transformation of ecol and ecol_HIS in pSB1C3 for plasmid isolation.
- Team Fungal and Plant Laccases: Plating positive colonies from cloning of tvel5 in pSb1C3 backbone.
- Team Site Directed Mutagenesis:
- Plasmid-isolation of two more ecol-g2307a colonies.
- Team Cellulose Binding Domain:
- Plated one colony of <partinfo>BBa_I13522</partinfo> on selection-agar.
- Dephosphorylation of pSB1C3 Backbone.
- Ligation of [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] with the pSB1C3-Backbone and [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)] with the pSB1C3-Backbone.
- Transformed both in KRX and plated on CM-selection-agar.
- Team Immobilization:
- CPC beads have just arrived! We couldn’t wait any longer to start our new experiments. Since the amount of beads used and the incubation period varied from one paper to another (see Literature), we decided to find out ourselves the most convenient conditions for our laccases. We started with different bead concentrations: 0.01, 0.02, 0.04, 0.06, and 0.08 mg/mL. The beads were first immersed in 2.5% glutaraldehyde and incubated for 2 hours under light vacuum in order to allow as much beads’ surface area to be coated with aldehyde groups, which crosslink the laccases to the beads. After that, the beads were washed 3 times with Britton-Robinson Buffer and then immersed in 1mg/mL laccases from TVEL0 and placed on a rotator at 4°C for 18h and 36h respectively. For further information see Protocol.
Tuesday August 7th
- Team Cloning of Bacterial Laccases:
- Plasmid isolation and control restriction of ecol and ecol_HIS in pSB1C3 showed correct fragment sizes in agarose gel. So we did a digest for prefix insertion of the new T7 promoter.
- Team Site Directed Mutagenesis:
- Digestion of the two ecol-g2307a-mutants showed that one has lost the restriction-site. Prepared that one for sequencing.
- Team Cellulose Binding Domain:
- Plasmid-isolation of <partinfo>Bba_I13522</partinfo>.
- PCR with <partinfo>BBa_I13522</partinfo> as template and GFP_Freiburg and GFP_His6_compl-primers and clean-up through agarose-gel.
- Restriktion of Linear Backbone, [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] and [http://partsregistry.org/Part:BBa_K863121 GFP_His] with EcoRI and PstI.
- Ligation of Linear Backbone with [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] and with [http://partsregistry.org/Part:BBa_K863121 GFP_His], respectively.
- Transformation of all that stuff and plating on selection-agar.
- Team Immobilization:
- The 18h samples (see above: Monday August 6th/Team Immobilization) were ready for further analysis. The supernatants were gathered to be tested for laccase activity. The beads were then washed with Britton-Robinson Buffer (link), then with 0.5M NaCl solution in order to wash away all noncovalently bound laccases and again twice with the buffer. Subsequently, the beads were immersed in 2.5 mg/mL glycine for 18h at 4°C.
Wednesday August 8th
- Team Shuttle Vector: Some more conditions were varied to get the fragment tAOX1 for the shuttle vector:
- An other annealing temperature gradient was run with 2% DMSO, 4% DMSO included samples and samples mixed with 5x GC buffer instead of 5x HF buffer. The right products were detected under the following conditions:
- In the sample with 2% DMSO and an annealing temperature of 40.9 °C.
- In the sample with 4% DMSO and an annealing temperature of 40.3 °C, 57.1 °C, 59.9 °C.
- In the sample with 5x GC buffer and an annealing temperature of 50.7 °C, 54.0 °C, 59.9 °C.
- Team Cloning of Bacterial Laccases:
- We dephosphorylated the digested the plasmids from day before and phosphorylated the promoter parts. After that we ligated the two parts and transformated the products into KRX electrocompetent cells.
- Team Fungal and Plant Laccases: Plasmid isolation of tvel5 laccase in pSB1C3 backbone.
- Team Site Directed Mutagenesis:
- Made PCRs on tvel-t143g-mutants with Tv_lac10.P.FW and Tv_lac10.S.RV primers, with products of 1.6 kbp when there should be about 4.0 kbp.
- Team Cellulose Binding Domain:
- Picked one not-red colony of [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)], [http://partsregistry.org/Part:BBa_K863112 CBDclos] and [http://partsregistry.org/Part:BBa_K863121 GFP_His] and plated it on seletion-agar
- Restriktion of [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)]-PCR-product with AgeI and EcoRI with DpnI
- Restriktion of [http://partsregistry.org/Part:BBa_K863121 GFP_His]-PCR-product with NgoMIV and PstI and DpnI
- Freiburg-Assembly with [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] and [http://partsregistry.org/Part:BBa_K863121 GFP_His] on the linearized pSB1C3 Backbone followed by plating on selection-agar.
- Team Activity Tests: Today was an exciting day for us. We got samples from Team Immobilization for activity measurements. They were trying to analyze which concentration of beads is a good choice for immobilizing the laccases. The exciting part was to check all the solvents they are using for possible reaction with ABTS during measurements. Check Team Immobilization´s Labjournal and protocols for the output and further information.
- Team Immobilization:
- The 18h samples (see above: Monday August 6th/Team Immobilization and Tuesday August 7th/Team Immobilization) were washed with Britton-Robinson Buffer (link), then with 0.5M NaCl solution and again twice with the buffer. The 36h samples were treated similarly as the 18h samples (see above: Tuesday August 7th/Team Immobilization). The supernatants were delivered to the “Activity Test” team to measure the activity of the non-bound laccases. Based on the results, we would be able to figure out the most convenient bead concentration and incubation period. After comparing the results, we found out that the activity of the supernatant after 36h was less than after 18h, which indicated that 36h incubation is more convenient. On the other hand, the activity also decreased upon increasing the beads’ concentration without reaching a saturation level (see Fig.). Therefore, we decided to carry out another experiment with higher beads concentration.
Thursday August 9th
- Team Cloning of Bacterial Laccases:
 Figure 1: Agarosegel from the Streptomyces gradient PCR. 1-12: S. goettingen in Sequence with following Annealing temp. 54.9°C, 55.1°C, 55.8°C, 56.8°C, 58.1°C, 59.5°C, 61.0°C, 62.4°C, 63.7°C, 64.8°C, 65.6°C, 66°C. 13-22: S. tuebingen in Sequence with following Annealing temp. 54.9°C, 55.1°C, 55.8°C, 56.8°C, 58.1°C, 59.5°C, 61.0°C, 62.4°C, 63.7°C, 64.8°C
Figure 1: Agarosegel from the Streptomyces gradient PCR. 1-12: S. goettingen in Sequence with following Annealing temp. 54.9°C, 55.1°C, 55.8°C, 56.8°C, 58.1°C, 59.5°C, 61.0°C, 62.4°C, 63.7°C, 64.8°C, 65.6°C, 66°C. 13-22: S. tuebingen in Sequence with following Annealing temp. 54.9°C, 55.1°C, 55.8°C, 56.8°C, 58.1°C, 59.5°C, 61.0°C, 62.4°C, 63.7°C, 64.8°C- We sent ecol and ecol_His (both in pSB1C3) for sequencing.
- We did again a gradient PCR for the Streptomyces but this time without S. tuebingen because the DNA we got was empty. The results can be seen on Figure 1.
- Team Fungal and Plant Laccases:
- Again: Ligation of tvel35 in pSB1C3 backbone.
- Plasmid isolation of tvel5 + pSb1C3 and control digest with NotI. For one of the two plasmids the digest looks good. We sent this plasmid for sequencing.
- Team Site Directed Mutagenesis:
- Sequencing results arrived:
- One bpul-plasmid is positive on both mutations
- All Xccl-mutants are negative (there even was a part of 900 bp gone missing!)
- plated six more colonies of the xccl-g3633c for plasmid-isolation
- PCR with the original tvel10-plasmid and the Prefix/Suffix-primers, showed no product at all.
- Sequencing results arrived:
- Team Cellulose Binding Domain:
- Plasmid-isolation of [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)], [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] and [http://partsregistry.org/Part:BBa_K863121 GFP_His].
- Transformated the assembly again with more of volume of the assembly-CBDcex-GFP_His-mix.
- Team Cultivation & Purification:
- We got E. coli KRX with our own BioBrick [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] in pSB1C3 like the rest.
- We discussed if we did not produce anything because of the toxicity of our protein, which may reduce the plasmid stability. We searched for maximal used concentration of chloramphenicol and found out that concentrations up to 170 µg/mL were used. Therefore we decided to start a new flask cultivation with concentrations of chloramphenicol varying in 5 steps from 20 µg/mL to 170 µg/mL. Today we made the precultures of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] and E. coli KRX as negative control.
Friday August 10th
- Team Cloning of Bacterial Laccases:
- Again we did the digest of our new T7 promoter part and the ligation in pSB1C3 backbone with ecol PCR products with and without HIS tag. After that we transformed the ligations in pSB1C3. Additionally we did the same with promoter J23110 instead of T7 promoter.
- We did PCR on [http://partsregistry.org/Part:BBa_K863020 BBa_K863020] with the primers B.halo_FW and B.halo_RV for cloning the gene in pSB1C3 backbone without promoter and HIS tag.
- We ligated the digested pSB1C3 plasmids with ecol and ecol_HIS with the new pT7 promoter and pSB1C3 backbone and transformed the approaches in KRX.
- Team Site Directed Mutagenesis:
- Plasmid-isolation xccl-g3633c-colonies and test-digestion showed no positive mutated plasmids.
- New pfu-PCR on the xccl-plasmid with xccl-g3633c primer-mix
- Team Cellulose Binding Domain:
- Picked [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His] assembly colony and plated it on CM-selection-agar for plasmid-isolation.
- Restricted [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)]with EcoRI and AgeI
- Digested insert did not appear in the
- Colony-PCR of [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)].
- Picked and plated several colonies of the [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His]-assembly and plated the on CM-Agar.
- Team Cultivation & Purification:
- Flask cultivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863005] to test various concentrations of chloramphenicol. We used E. coli KRX as negative control.
- Settings: 100 mL flasks without baffles, final volume: 10 mL, autoinduction medium with 20/ 57.5/ 95/ 132.5/ 170 µg/mL chloramphenicol, 37 °C, 120 rpm, double determination.
- Note: Found out, that we did not use the maximal possible concentration of chloramphenicol. We decided to use a higher concentration than 20 µg/mL and chose 60 µg/mL for further cultivations. This is a compromise between benefits and costs.
- Flask cultivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863005] to test various concentrations of chloramphenicol. We used E. coli KRX as negative control.
Saturday August 11th
- Team Cloning of Bacterial Laccases:
- Colony PCRs showed positive colonies for ecol(J23110) so we plated this colonies on new plates.
- We did the PCRs of the laccase genes ecol, bpul, bhal and lthl again. We used the …_FW / …_RV primers and the …_FW / …_RV_HIS primers of the different genes. Digestion of this PCR products and ligation with pT7 or promoter J23110 and the pSB1C3 plasmid backbone.
- Team Site Directed Mutagenesis:
- Agarose-gel-electrophoresis of xccl-PCR (yesterday) showed bands at 3.2 kbp and over 12 kbp but not at 4 kbp as it should be.
- Team Cellulose Binding Domain:
- A few of the plated pSB1C3+CDBcex+GFP-assembly colonies have grown
- Colony-PCR [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)]-colonies showed no positive clones.
Sunday August 12th
- Team Cloning of Bacterial Laccases:
- PCR cleanup of the PCR products from the day before.
- We did control digest and got bands for ecol with J23110 promoter and with the new pT7. So we sent this plasmids for sequencing.
- Team Site Directed Mutagenesis:
- pfu-PCR with xccl-plasmid and xccl-g2247c primers resulted in no product.
- PCR with pre- and suffix primers for tvel10 with original Topo-plasmid as template with increasing temperature at each step (0.1°C) resulted in no product.
- Team Cellulose Binding Domain:
- Colony-PCR for [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)], [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)] and [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His].
- resulted in two positve [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)]-colonies.
- Did another colony-pcr on [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] and [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His] that brought up nine positive clones of [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)].
- Colony-PCR for [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)], [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)] and [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His].
- Team Activity Tests: Today was another day for finding the mistake. Team Cultivation used different methods the for lysis of E.coli KRX cell extract to check whether our [http://partsregistry.org/Part:BBa_K863005 ECOL] laccases and thus their activity might me damaged during lysis. Compared were an enzymatic lysis via lysozyme and a chemical lysis via B.Per/DNase, also both ways were combined in the third sample We are sorry to tell that no activity was seen.
Summary of Week 16
hier eine Zusammenfassung
Week 16 (08/13 - 08/19/12)
Contents |
Weekly Seminar
- As part of our collaboration the iGEM team from SDU-Denmark will arrive on saturday (25th of august).
- Our StreetScience booth will have 16 m2.
- We already ordered all info-material we want to provide at StreetScience.
- Just in case we are going to qualify for the world finals in Boston, everyone needs a visa. Especially our team members without a european nationality have to inform about visa terms.
- Derya is responsible for planning a new, project-driven video.
- As deadlines come closer we have to think about a team name.
- He have decided to rent a bus to travel to Amsterdam.
Monday August 13th
- Team Cloning of Bacterial Laccases:
- After cleaning up the gradient PCR from the 09th August we did an digestion with the S. goettingen DNA for cloning it into the pSB1C3. The pSB1C3 was additionaly treated with Shrimp-alkaline-phosphotase. After this we set the ligation of insert DNA and vector DNA.
- Team Site Directed Mutagenesis:
- Gradient-PCR of tvel10 55 to 66°C with DMSO (12 Steps) resulted in a lot of product at 55°C and 63°C to 66°C; a little product at 56°-59° and 61°-62°C and no product at 60°C (59°C was the temperature Clonemanager predicted and I used before). Merged the products and cleaned them up.
- Digested the product with EcoRI, PstI and DpnI.
- Team Cellulose Binding Domain:
- We did another PCR on the <partinfo>I13522</partinfo> with the [http://partsregistry.org/Part:BBa_K863121 GFP_His]-primers with positive result.
- Plasmid-isolation of two positive clones of [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)] followed by digestion with NotI that confirmed that the plasmid is [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)]. Prepared both for sequencing.
- Plated three positve [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)]-colonies.
- Restriction of the [http://partsregistry.org/Part:BBa_K863121 GFP_His]-PCR-product with EcoRI and PstI to bring it into the pSB1C3-backbone.
- Also: Restriction of the [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)]-PCR-product with NgoMIV and PstI for an assembly.
- Restriktion of the [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)]-PCR-product with AgeI and EcoRI for the assembly.
- All digestions were done with DpnI to get rid of the templates.
- Ligation of [http://partsregistry.org/Part:BBa_K863121 GFP_His] with the pSB1C3-backbone.
- Ligation of [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7) and GFP_His] with the pSB1C3-backbone.
- Transformed all ligations into KRX.
- Team Cultivation & Purification:
- Today we discussed that we may get activity if we start to purify our laccases. So from now we will purify our products before measuring them. We hope this will bring promising results. Therefore we searched for a useful method.
Tuesday August 14th
- Team Cloning of Bacterial Laccases: We finally had positive bacteria for the S. goettingen. So we decide to do a colony PCR. But the result has shown that there were no insert DNA but we didn't want to give it up so we transfered some colonies to a new plate an let them grown over night again.
- Team Fungal and Plant Laccase: Today our cDNA took a bath in ethanol and got cleaned. We are pretty sure our next PCR will be a lot more successful. Check protocols for further information about the cDNA washing via ethanol precipitation.
- Team Cultivation & Purification:
- Made preculture of E. coli KRX without plasmid and with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] as well as [http://partsregistry.org/Part:BBa_K525710 BBa_K525710].
- Team Site Directed Mutagenesis:
- Ligation of tvel (PCR-product) and pSB1C3 and transformation into KRX
- Gradient-PCRs (55°C bis 72 °C) with xccl-plasmid using xccl-g3633c and xccl-g2247c primer-mixes, respectively, resulted in no product of the right size.
- Team Cellulose Binding Domain:
- Sequencing results showed, that the insertion of [http://partsregistry.org/Part:BBa_K863121 GFP_His] wasn't successful at all, [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] had a lot of mutations and [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)] had one silent mutation.
- On the [http://partsregistry.org/Part:BBa_K863121 GFP_His]-transformation-dish did not grow any colony. Did a new transformation with a little more ligation-product and plated the cells on selection-agar.
- Plasmid-isolation of the three positive [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)]-plasmids and digestion showed not the correct fragments. Plated some more positive colonies on selection-agar for plasmid-isolation.
- The [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His]-dish had only nine colonies and a colony-PCR showed that none of them had the right insert.
- Team Activity Tests:
- Again our skills in measuring laccase activity were in demand today. We had a job incoming from Team Substrate Analytic. Since they need to dissolve their substrates in methanol or acetonitrile, they were curious about laccase activity related to these solvents. And they infected us with their curiosity! So we started measurements using 0.1 U TVEL0 Laccase, 40 µL sodium acetate buffer and different amounts of MeOH or acetonitrile respectively, ad 200 µL H2O. Our tested range went from 2 µL to 18 µL. The results show that MeOH and acetonitril have an impact on the ability of TVEL0 to oxidize ABTS. Using 16 µL MeOH (which means 8% of the amount of the sample contained MeOH) led to a different saturation curve as usual. The saturation couldn't be reached any more (see Fig. 1). Acetonitrile had a noticeable impact when using 5% (14 µL) of it in relation to the sample (see Fig. 2). But in the end both solvents couldn't stop TVEL0 laccase from being active. We are happy to tell Team Substrate Analytics that they can proceed as planned and dissolve the substrates in the tested solvents.
- Team Substrate Analytic:
- Our first attempt do detect diverse estrogens with the clairity HPLC. Dominic helped us to set the HPLC method based on the paper „Enzyme-Catalyzed Oxidation of 17β-Estradiol Using Immobilized Laccase from Trametes versicolor”.
- We prepared dilution series from three different estrogens and tryed to mesure them. Sadly all substrates left the column with the same retention time as the solvent.
Wednesday August 15th
- Team Wiki: We set up some more wiki rules. Today´s rules were about citations. We agreed about some standards that will for sure help make the wiki a little prettier. Have a look:
- Paper: Exampleman M et al. (2002). The example paper. The example journal (Volume): pages.
- Website: Name of website, URL, Index, date site visited
- Book: Examplewomen M et al. (1999). The example book. The example publisher (edition).
Next will be a standard for charts.
- Team Cloning of Bacterial Laccases:
- Plasmid isolation from the plate of the 14th August. This was done with the Analytic Jena Kit.
- Team Site Directed Mutagenesis:
- Colony-PCR of tvel10-colonies resulted in small bands. Quickly explained: tvel10 still had illegal PstI-restrictions-sites. Digestion of tvel10 PCR-product with NotI as well as digestion of RFP-pSB1C3 with NotI
- Team Cellulose Binding Domain:
- Plasmid-isolation of the three positive [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)]-colonies, but digestion showed no positive result, again.
- Colony-PCR of [http://partsregistry.org/Part:BBa_K863121 GFP_His] and [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His] did not show any positive result, ether.
- Team Cultivation & Purification:
- Flask cutivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000],[http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015]. We used E. coli KRX negative control as well as E. coli KRX with [http://partsregistry.org/Part:BBa_K525710 BBa_K525710] as positive control.
- Settings: 1 L flasks without baffles, final volume: 250 mL, autoinduction medium supplemented with 60 µg/mL chloramphenicol, 37 °C, 120 rpm, single determination
- Flask cutivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000],[http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015]. We used E. coli KRX negative control as well as E. coli KRX with [http://partsregistry.org/Part:BBa_K525710 BBa_K525710] as positive control.
- Team Activty Tests: Today we got another mission from Team Cultivation. They gave us a huge amount of laccase samples for measurements. The samples contained as well general laccase samples for activity tests but also the flow through from the purification process to check if any laccase is lost during this process. The results show no enzyme activity in any case.
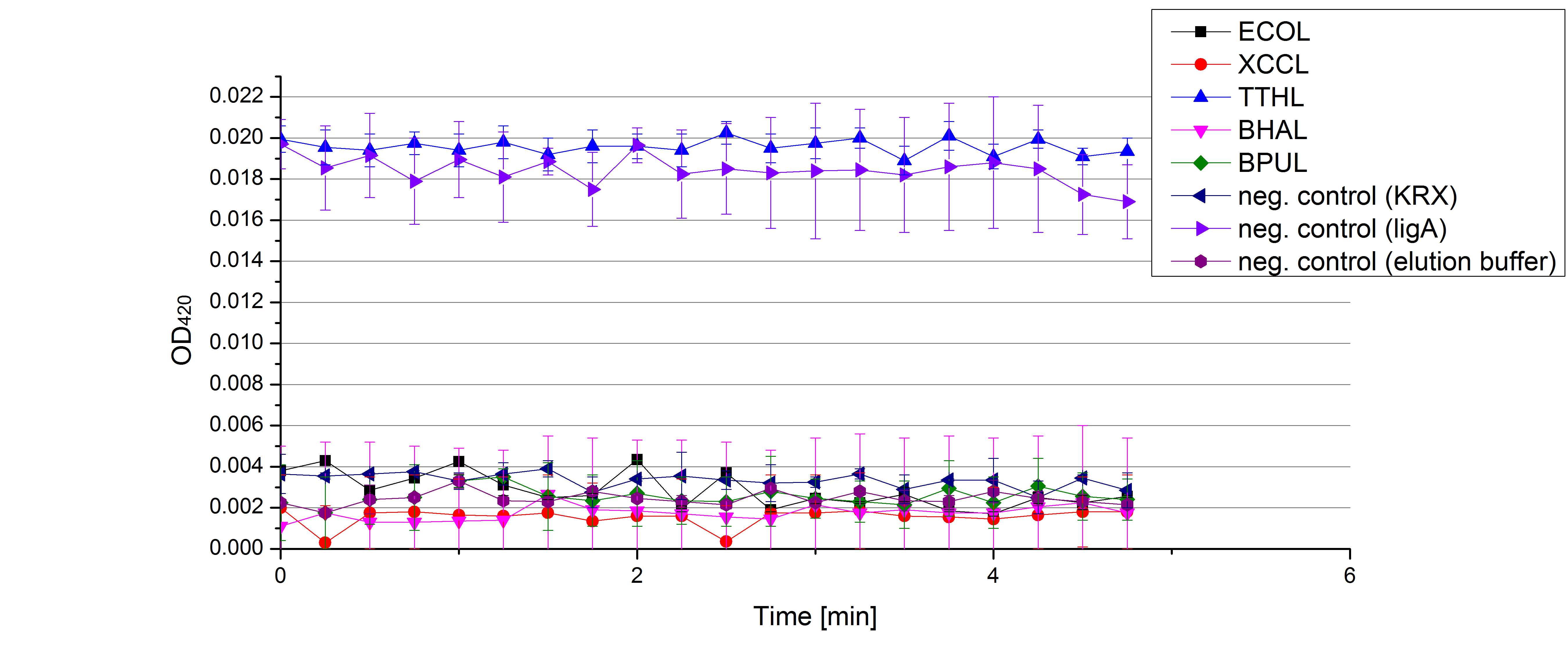
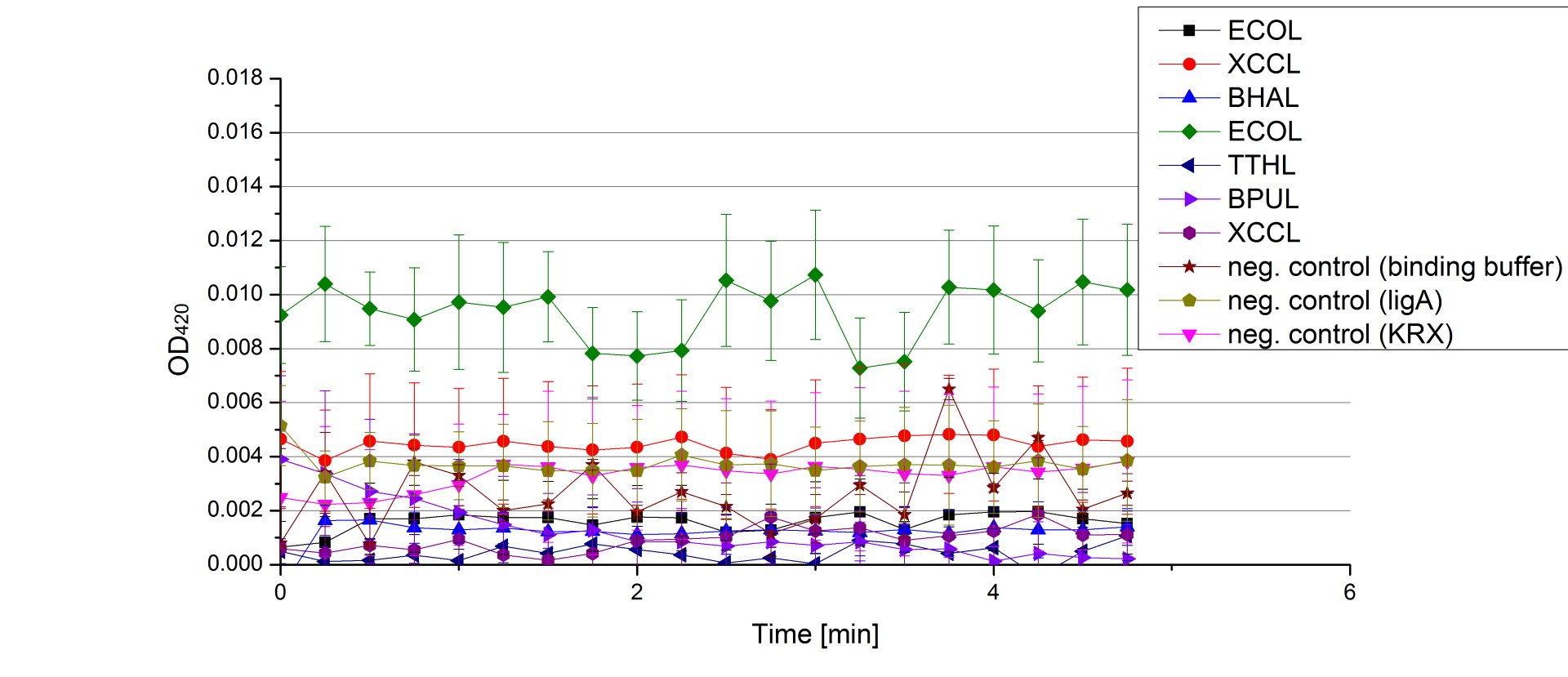
- Team Substrate Analytic:
- Dominik tried to regenerate the column. After that we measured new dilution series from all three estrogens. Once again all substrates left the column together with the solvent. That means the column is defect.
- We ordered new C18-columns from Knauer and waited for their arriving. A Eurosphere and a Bluespehere column.
Thursday August 16th
- Team Shuttle Vector: For the Gibson assembly we need more higher concentrated fragments. In fact, Phusion PCR was done for the fragments 5AOX1, 3AOX1, Kozak-MFalpha1 and pSB1C3 at following conditions with 35 cycles. PCR fragments were used as DNA template.
| Step | Condition |
|---|---|
| Initial denaturation | 98 °C, 30 s |
| Denaturation | 98 °C, 10 s |
| Primer annealing | 67 °C, 20 s |
| Elongation | 72 °C, 90 s |
| Final Elongation | 72 °C, 5 min |
Phusion PCR was also done for the HIS4 fragment at following conditions with 35 cycles and also PCR product as template.
| Step | Condition |
|---|---|
| Initial denaturation | 98 °C, 30 s |
| Denaturation | 98 °C, 10 s |
| Primer annealing | 60 °C, 20 s |
| Elongation | 72 °C, 90 s |
| Final Elongation | 72 °C, 5 min |
Phusion PCR was also done for the tAOX1 fragment at following conditions with 35 cycles and also PCR product as template. For us it can not be recognized at which annealing temperature the primer binds best. That is why we choose an annealing temperature gradient more. This time from 50 °C until 60 °C. The mixture of the sample is the same as last time.
- Team Site Directed Mutagenesis:
- Clean-up of tvel10 and pSB1C3 (both cut with NotI); Dephosphorilation of pSB1C3 with SAP and ligation. Transformed into KRX and plated on select-agar.
- Team Cellulose Binding Domain:
- Colony-PCR of [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His]-clones with no result, even the positive-control was negative, so we plated three colonies for a test-digestion instead.
- Digestion of [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)] with AgeI and PstI and digestion of the [http://partsregistry.org/Part:BBa_K863121 GFP_His]-PCR-product with NgoMIV and PstI over-night.
- Team Cultivation & Purification:
- Today we harvested the cells from the cultivation on 08/15, disrupted them in equilibration buffer via sonication, centrifugated and purified the supernatant via HisTrap column. The purified samples in 500 mM Imidazol were given to the activity test team.
Friday August 17th
- Team Cellulose Binding Domain:
- Stopped digestion of [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)] and [http://partsregistry.org/Part:BBa_K863121 GFP_His]. Dephosphorylated pSB1C3+[http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)].
- Plasmid-isolation of the three [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His]-dishes.
- The sequencing of the two [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)]-plasmids showed the same result as the first: one silent mutation. We decided to carry on with this plasmid.
Saturday August 18th
- Team Cloning of Bacterial laccases:
- Today we performed a ligation from the digestions we have done before. The ligation included of course the T7 promotor, the backbone pSB1C3 and the individual laccase genes. Ligation was performed with T4 ligase for 20 minutes at room temperature and stopped through a five minute incubation at 70 °C.
- Team Cellulose Binding Domain:
- Ligated [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)] and [http://partsregistry.org/Part:BBa_K863121 GFP_His], transform into KRX and plated on CM-selection-Agar.
Sunday August 19th
- Team Site Directed Mutagenesis:
- Colony-PCR of tvel10-colonies with three positive result.
- Plated those three on select-agar for plasmid-isolation.
- Team Cellulose Binding Domain:
- Colony-PCR of colonies from the [http://partsregistry.org/Part:BBa_K863113 CBDclos(T7)+GFP_His]-trafo-dish with only negative results.
Summary of Week 17
hier eine Zusammenfassung
Week 17 (08/20 - 08/26/12)
Contents |
Weekly Seminar
- Our denish guests from SDU-Denmark will stay from 25th to 27th of august.
- Program will be as follows: On saturday together we will present our teams on StreetScience and saturday evening it is time for Eurodance with music from David Hasselhoff. On sunday we will have breakfast together and after we will have a scientific symposium with discussing our projects, problems and trouble shooting. On sunday evening we will show our denish guests the cozy inner part of Bielefeld.
- StreetScience: we can't rent televisions from Bielefeld Marketing, but Derya could organize two other big televisions. We will have a light microscope and our dress code will be to wear a black t-shirt.
- Agatha and Saskia will bring Volvox, Nadine will bring Xanthomonas and Robert Corynebaterium. All strains are harmless and wild-type strains.
- Mo organizes some games for kids and Agatha&Saskia will tinker prices.
- Start of preparation is 9.30
Monday August 20th
- Team Shuttle Vector: A Gibson assembly was done as described in the protocol, but only 10 ng could be used, because the concentration of the fragments was to low. The sample was transformed in E. coli KRX via electroporation and plated out on selective LB Cm plates.
- Team Cloning of Bacterial Laccases:
- Digestion of pSB1C3 plasmid backbone, tthl_HIS and bpul_HIS for doing the assembly with the new T7 promoter and J23110 again.
- We did PCRs on bpul_HIS, bhal_HIS and ecol_HIS again.
- Team Cellulose Binding Domain:
- Biggg Colony-PCR of the [http://partsregistry.org/Part:BBa_K863113 CBDclos(T7)+GFP_His]-ligation with the result, that all seem to carry [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)]. The ligation didn't work at all...
- Team Cultivation & Purification:
- Today we decided to finish our lean period of non-active laccases. It was time for a transcript analysis to have a look if our laccases are transcribed into RNA and if there is a lot of it or if its gets degraded just in time. We took our cells, centrifuged and shock freezed them in liquid nitrogen to stop every metabolism in the cell and freeze the status of the RNA. The frozen cell pellets were stored at -80°C for the upcoming tests. As well we designed and ordered primers.
- Team Immobilization
- Today we set up a new immobilization experiment following the same procedure as in week 15 (see week 15 Monday August 6th-Wednesday August 8th /Team Immobilization) but with different bead concentrations: 0.06, 0.08, 0.1, 0.12, 0.14 and 0.16 mg/mL. Based on the results of the first experiment, the samples were incubated with TVEL0 for 36h.
Tuesday August 21st
- Team Shuttle Vector:
- Design of primers for the Part his4 [http://partsregistry.org/Part:BBa_K863202 K863202].
- Colonies from yesterday were plates out and Colony-PCR was done with GoTaq and the F and R primer to amplificate the part [http://partsregistry.org/Part:BBa_K863204 K863204].
- From positive clones the plasmid was isolated and restriction analyses with NotI was done. No one of the clones has passed the restriction analyse.
- Team Cloning of Bacterial Laccases:
- The isolated plasmid from the 15th August was prepared and given up for sequencing.
- We designed primers for q-PCR for Team Activity Tests (details, look above). The PCRs with this primer pairs should result in about 200 bases long fragments.
- We realized that the primer pairs we anneal for the promoter parts had about 2000 ng/µl if diluted 1:10 from originally 100 pmol. Maybe our used amounts on promoter for the assemblies was a LITTLE to high. So now we try a dilution of 1:1000 with about 2 ng/µl.
- PCR products of different laccases were purificated and digested for suffix insertion.
- Ligation of the laccase genes ecol, ecol_HIS and bpul and bpul_HIS, the 1:000 diluted (0,1 pm/µl) promoters (pT7 and P110) in pSB1C3 vector.
- Ligation of the digested ecol_pSB1C3 and ecol_HIS_pSB1C3 plasmids with the 1:000 diluted (0,1 pm/µl) promoters (pT7 and P110).
- Ligation of T7 promoter and J23110 promoter in pSB1C3 backbone.
- Team Fungal and Plant Laccases:
- We designed primers for the laccase <partinfo>BBa_K500002</partinfo> for cloning in P. pastoris shuttle vector.
- Team Cellulose Binding Domain:
- Tested restriction enzymes.
Wednesday August 22nd
- Street Science:
- Went to Dr. Joe Max Risse today and asked him about the mircoorganisms of the Fermentationgroup. He recommended Euglena gracilis, since it is without risk, big, colorful, and very healthy under the microscop.
- I asked for the bacteria they have and he told me I could check if their Kocuria rosea is a wild type.
- In the DMSZ catalog there are three strains of Kocuria rosea and all have been assessed to be of riskgroup 1, which is the safest group of bacteria and means it is unlikely that it will infect humans.
- I also asked for Penicillium chrysogenum but the one they use is a GVO.
- Team Cloning of Bacterial Laccases:
- Colony PCRs: some prefix insertions of promoters in pSB1C3 with ecol and ecol_HIS showed positive bands, so we picked the colonies for plasmid isolation. The problem with the prefix insertion of the promoters in pSB1C3 with the ecol gene is that we can’t easily prove that the promoters were ligated in the backbone because with control restriction there would be an about 50 bp longer fragment which is difficult to see even in 3 % agarose gel. For this reason we designed FW primers (J23103_K_FW and J23110/117_K_FW and T7_K_FW) for our colony PCR, which can just bind, if the new promoter sequence is included in the plasmid. Until all colonies from the promoter J23110 ligation were red, we had some white colonies for the pT7 promoter in pSB1C3 backbone. So we picked a positive colony and plated it for plasmid isolation.
- We plated colonies on new plates, which weren't red from the transformation of pSB1C3 + pT7. We can't do colony PCR on this plasmids, because the resulting parts would be smaller than 100 bps.
- Team Site Directed Mutagenesis:
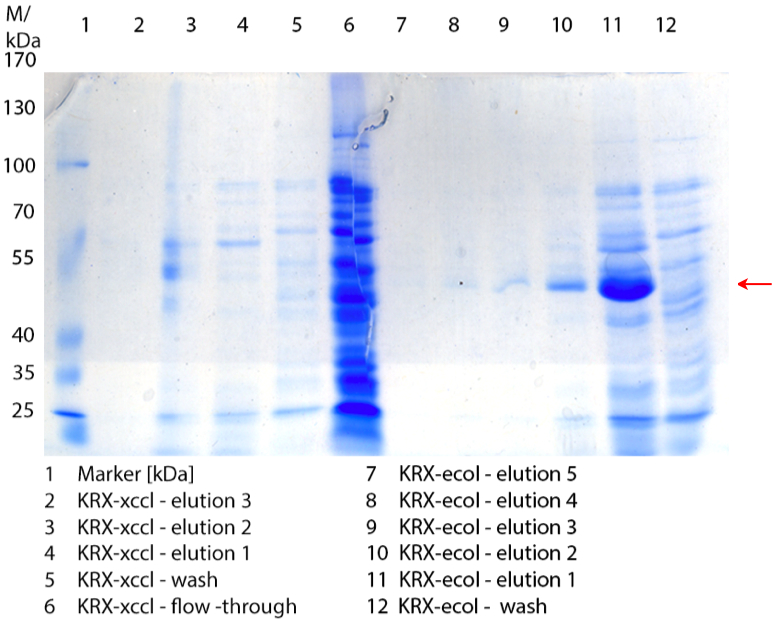 Figure 1: SDS-PAGE of purification from cultivation (08/15) of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000],[http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015], E. coli KRX (negative control) and E. coli KRX with [http://partsregistry.org/Part:BBa_K525710 BBa_K525710] (positive control). 250 mL in 1 L flasks without baffles, autoinduction medium with 60 µg mL-1 chloramphenicol at 37 °C for 12 hours. Purification of the supernatant via syringe method. The arrow shows ECOL in lane 10 and 11 (elution 1 and 2).
Figure 1: SDS-PAGE of purification from cultivation (08/15) of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000],[http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015], E. coli KRX (negative control) and E. coli KRX with [http://partsregistry.org/Part:BBa_K525710 BBa_K525710] (positive control). 250 mL in 1 L flasks without baffles, autoinduction medium with 60 µg mL-1 chloramphenicol at 37 °C for 12 hours. Purification of the supernatant via syringe method. The arrow shows ECOL in lane 10 and 11 (elution 1 and 2).
- Ordered new primers for Xantomonas Campestris SDM, since there is still no positive colony and even gradient-PCR gave the wrong product
- Team Cellulose Binding Domain:
- Test-restriction of all isolated [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)]- and [http://partsregistry.org/Part:BBa_K863121 GFP_His]-plasmids with NotI with no positive result.
- Restarted from the beginning and made a gradient-PCR for [http://partsregistry.org/Part:BBa_K863121 GFP_His] and [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] to get some more product.
- Team Cultivation & Purification:
- Made SDS-PAGES of cultivation from 08/15.
Thursday August 23rd
- Team Cloning of Bacterial Laccases:
- Made plasmid isolation of pSB1C3 + pT7 and sent plasmids for sequencing.
- Team Fungal Laccases: We did PCRs on tvel35 plasmid with primer pairs Pc_lac35.P.FW / Pc_lac35.S.RV for trying ligation in pSB1C3 again and with Pc_lac35_FW_oS / Pc_lac35_RV for cloning in shuttle vector.
- Team Cellulose Binding Domain:
- Clean-up of [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)]- and [http://partsregistry.org/Part:BBa_K863121 GFP_His]-PCR-product (pooled good ones and bad ones, respectively).
- Restriktion of the [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)]-PCR-product with EcoRI and PstI for ligation with the backbone and EcoRI and AgeI for a assembly.
- Restriktion of the [http://partsregistry.org/Part:BBa_K863121 GFP_His]-PCR-product with EcoRI and PstI for ligation with the backbone and NgoMIV and PstI.
Friday August 24th
- Team Site Directed Mutagenesis:
- Plasmid-isolation and digestion of tvel10 colonies with NotI showed that two of the plasmids have an insert of the right size.
- Team Cellulose Binding Domain:
- Stopped restrictions and clean-up them up. Dephosphorilated the linearized backbone and ligated it to [http://partsregistry.org/Part:BBa_K863121 GFP_His], [http://partsregistry.org/Part:BBa_K863121 GFP_His] and assembled [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His].
- [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)]-cells plated to get some more plasmid.
Saturday August 25th
STREET SCIENCE Visit of Danish guys till 08/27
Sunday August 26th
- Team Cellulose Binding Domain:
- Made-up the idea to have a constitutive promoter (like <partinfo>BBa_J61101</partinfo>) in front of the cellulose binding domain and the GFP. With a Freiburg-Prefix in front of the CBDs we could even easily switch directions of CBD and GFP.
- Transformed <partinfo>BBa_J61101</partinfo> from the iGEM-plate into KRX and plated it on AMP-selection-dish.
- Team Cultivation & Purification:
- Made precultures of E. coli KRX without plasmid (negative control) and with plasmids containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015]. As postive control we used [http://partsregistry.org/Part:BBa_K525710 BBa_K525710 (Ligase A)].
Summary of Week 18
hier eine Zusammenfassung
Week 18 (08/27 - 09/02/12)
Contents |
Weekly Seminar
Summary of the weekend with our denish guests
- We will try to collaborate in modelling and Julia V tries to help the iGEM team from SDU-Denmark to build up the wiki.
Summary of StreetScience
- Our commonday SynBioDay - StreetScience in Bielefeld was success, although it was a little chaotic in the beginning.
- Derya will evaluate our feedback poll.
Deadlines
- Mo is responsible for our registration to the jamboree.
- Miriam is responsible for our safety page. She has nearly finished the texts so everyone has to review the page.
- Gabi and Robert will write the abstract.
- Next week René Röspel, member of the german parliament, will visit us for a discussion about iGEM and Synthetic Biology.
Monday August 27th
- Team Cloning of Bacterial Laccases:
- Digest of our new plasmid pSB1C3 with pT7 promoter (hopefully, cause we have to wait for sequencing results). Dephosphorylation of digested vector and purification over agarose gel. After that, the backbone was ligated with ecol_HIS, tthl_HIS, bpul_HIS and bhal_HIS inserts.
- Again ligation of J231110 in pSB1C3 backbone.
- Team Cellulose Binding Domain:
- Colony-PCR [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)], [http://partsregistry.org/Part:BBa_K863121 GFP_His] and [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His] with no positive result at all.
- Restriction of pSB1C3+[http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His] with PstI and AgeI followed by dephosphorylation.
- Restriction of pSB1C3+RFP with XbaI and PstI, followed by dephosphorylation.
- Restriction of [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)]-PCR-product with XbaI and PstI
- Restriction of [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)] and [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)]-PCR-products with XbaI and AgeI
- Transformed the [http://partsregistry.org/Part:BBa_K863113 CBDclos(T7)+GFP_His]-ligation into KRX and plated cell on selection-agar.
- Team Site Directed Mutagenesis:
- Digestion of two tvel10-plasmids with BamHI and PstI to define direction of the insert. One is positive, one negative.
- Made the positive tvel10-plasmid ready for sequencing.
- Team Cultivation & Purification:
- We exchanged the buffer of the purified ECOL from 08/14, which showed a promising band in the SDS-Page.
- Made a flask cultivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] or [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020] with positive ([http://partsregistry.org/Part:BBa_K525710 BBa_K525710]) and negative control (without plasmid).
- Settings: 1 L flask without baffles, autoinduction medium, final volume: 250 mL, 60 µg/mL chloramphenicol, 37 °C, 120 rpm, durance: 12 hours, single determination
- Cells from today's cultivation were disrupted via sonication and laccase was purified by using the HisTrap column.
- Made precultures of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] or [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010]. As negative control we used E. coli KRX and as positive control we used E.coli KRX with [http://partsregistry.org/Part:BBa_K525710 BBa_K525710].
- Team Activity Tests:
- There have been some discussions about the storage of our laccases. It was said that the freezing of laccase (once deluted) might cause increased enzymatic activity. To find out whether storing the enzymes in the refrigerator or in the freezer is a better choice we compared to different samples from TVEL0. One was stored cooled and one was frozen and then measured as usual. As shown in figure 1 the frozen laccases showed an increased saturation curve and also reached the saturation slower. This seems like a clear hint to better store our enzymes in the refrigerator.
Tuesday August 28th
- Team Cloning of Bacterial Laccases:
- Ligation of pSB1C3 with promoters J23117, J23110 and J23103. The promoters (which are again primer pairs we annealed) were diluted 1:1000 (0,1 pmol) before and boiled with 98°C and slowly cooled down for ligation of the forward and the reverse primers.
- Colony PCR of ligated products with T7 promoter and insert. The PCR showed for the tthl_HIS and for ecol a band in agarose gel on the correct height. So we picked this colonies and plated them to isolate the plasmids.
- Team Cellulose Binding Domain:
- Gel-clean up of the digested pSB1C3-backbone.
- Colony-PCR of [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] and [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His] with four positive colonies of [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)].
- Plasmid-isolation of one [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His] followed by restriction with no positive result.
- Plated another [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His]-colony for plasmid-isolation.
- Plated small dots of 25 colonies of the [http://partsregistry.org/Part:BBa_K863113 CBDclos(T7)+GFP_His]-transformation to look if they are red.
- Plasmid isolation of <partinfo>J61101</partinfo> and digestion with SpeI and PstI
- Team Fungal and Plant Laccases:
- Digest of tvel35 PCR product for cloning in pSB1C3.
- Team Cultivation & Purification:
- The band appearing in the SDS-PAGE of the cultivation of the 08/14 was analysed via MALDI-TOF and we got a positive result: we got our laccase!
- We made a SDS-PAGE from yesterday's cultivation and got the same band for cultivation of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], so we reproduced our result:) It seems, that the purification and the higher temperature had the essential influence on the production.
- Started a new flask cultivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] or [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020] with positive ([http://partsregistry.org/Part:BBa_K525710 BBa_K525710]) and negative control (E. coli KRX without plasmid).
- Settings: 300 mL flasks without baffles, final volume: 60 mL, autoinduction medium, 0,35 mM CuCl2, 37 °C, 120 rpm, durance: 12 hours
- Cells from today's cultivation were disrupted via sonication and laccase was purified by using the HisTrap column. This time the column was better cleaned by using the twofold volume of 500 mM imidazol.
- Team Substrate Analytic:
- Dominik installed the new column into the HPLC.
- We tryed again to mesure the three estrogens,but we recived the same result as with the old column.
- We lowerd the oven temperature to 30 °C. With this settings we could determin the retention times 5.8 minutes for Estradiol, 4.7 minutes for Estrone and 5.2 minutes for Ethinyl estradiol.
Wednesday August 29th
- Team Cloning of Bacterial Laccases:
- Restriction of ecol_HIS, bpul_HIS, tthl_HIS and bhal_HIS PCR products estimated for the hundredth time.
- Team Cellulose Binding Domain:
- Plasmid-isolation of two plated [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)]- and the [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His]-coloniy and test-digestion of the plasmid with NotI showed fragments of the correct size.
- Colony-PCR of [http://partsregistry.org/Part:BBa_K863113 CBDclos(T7)+GFP_His]-colonies brought up no positive colonies.
- To have a comparison to our His-tagged-GFP-fusion-protein we decided to use an exact copy of the His-tagged-GFP. Therefore we made primers to add the His-tag the <partinfo>I13522</partinfo>.
- Made gradient-pcr with <partinfo>I13522</partinfo> as template and the GFP_FW and GFP_His_compl primers. the most correct product was at 62°C.
- Team Site Directed Mutagenesis:
- New Primers for xccl arrived.
- Team Cultivation & Purification:
- Made SDS-PAGES of yesterday's cultivation, but it was too pale, so it had to be repeated
- Made SDS-PAGES-repeat of yesterday's cultivation.
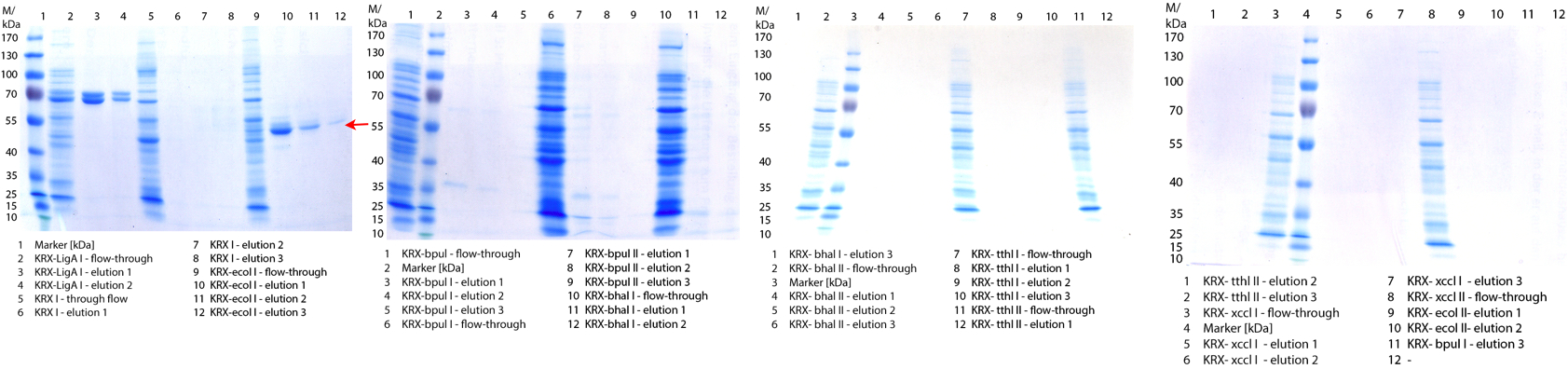
- Installation of the two 3 L fermenters used for practical courses in our lab. Had to search for all of the components with the help of members of fermentation group and test electrodes.
- Made precultures of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] or [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020] as well as of positive ([http://partsregistry.org/Part:BBa_K525710 BBa_K525710]) and negative control (E. coli KRX without plasmid).
- Team Substrate Analytic:
- Marcus Persicke has detected the Estrone and Estradiol limit.
- Dominic installed the arived columns from Knauer into the HPLC.
- We prepared samples in the same way as on the 14te August.
- But again, despite the new column all samples had the same retention time, as the solvent. So we reduced the oven tempreture to 30°C. With this settings we´ve got the 5min as retention time for E1 5,8min as retention time for E2 and 5,5 as retention time for EE.
Thursday August 30th
- Team Cloning of Bacterial Laccases:
- Ligation of the digested PCR products from the day before in BBa_J61101_pSB1A2 plasmid backbone which we transformed from distribution plate before.
- Ligation of J23110 and J23103 (annealed primers) in pSB1C3 backbone.
- Team Fungal and Plant Laccases:
- Digest of tvel35 again and ligation in pSB1C3.
- PCR of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K500002 BBa_K500002] with K500002_FW and K500002_RV primers.
- Team Cellulose Binding Domain:
- Pooled the six good and the six other PCR-fractions with producd of [http://partsregistry.org/Part:BBa_K863122 const.GFP_His]-gradient-PCR.
- Clean-Up of the PCR-products
- Start of Restriktion GFP-His6tag with EcoRI, PstI and DpnI in O-Buffer .
- Colony-PCRs of [http://partsregistry.org/Part:BBa_K863121 GFP_His] - All negativ.
- Pooled the six good and the six other PCR-fractions with producd of [http://partsregistry.org/Part:BBa_K863122 const.GFP_His]-gradient-PCR.
- Team Cultivation & Purification:
- Started another flask cultivation, the repetition of the cultivation on monday in a smaller scale but with a double determination. Cultivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] or [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020] with positive ([http://partsregistry.org/Part:BBa_K525710 BBa_K525710]) and negative control (E. coli KRX without plasmid).
- Settings: 300 mL flasks without baffles, autoinduction medium, final volume: 60 mL, 37 °C, 120 rpm, 12 hours
- Continuing Installation of the two 3 L fermenters.
- Starting the first fermentation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005]
- Settings: fermenter: Braun, autoinduction medium, final volume: 3 L, 37 °C, stirrer on cascade to hold a pO2 of 50 %, airflow: 2 NL/m, 12 hours. We had some problems with the controllation of settings.
- Made precultures of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] or [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020] as well as of positive ([http://partsregistry.org/Part:BBa_K525710 BBa_K525710]) and negative control (E. coli KRX without plasmid).
- Started another flask cultivation, the repetition of the cultivation on monday in a smaller scale but with a double determination. Cultivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] or [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020] with positive ([http://partsregistry.org/Part:BBa_K525710 BBa_K525710]) and negative control (E. coli KRX without plasmid).
- Team Substrate Analytic:
- 100 µg ml-1 and 10 µg ml-1 Estradiol is to high concentrated, to integrate the peak area. Concentrations around 1 µg ml-1 seems to be ideally for our HPLC.
Friday August 31th
- Team Cloning of Bacterial Laccases:
- Colony-PCRs of the ligations with J23100 + J61101 + ecol_HIS, bpul_HIS, tthl_HIS and bhal_HIS. Positive colonies for for ecol_HIS and tthl_HIS and bpul_HIS were plated on LB + AMP.
- We isolated plasmids with pSB1C3 + promoter J23117 from a former ligation and transformation.
- Team Fungal and Plant Laccases:
- Purification of PCR product [http://partsregistry.org/wiki/index.php?title=Part:BBa_K500002 BBa_K500002].
- Team Cellulose Binding Domain:
- Ligation of [http://partsregistry.org/Part:BBa_K863122 const.GFP_His] with a pSB1C3 and transformation of KRX with the product of the ligation.
- Team Site Directed Mutagenesis:
- sequencing of tvel10-plasmid showed a mutation at base 1573, altering the second last amino acid from threonine to asparagine
- Team Cultivation & Purification:
- Cells of the yesterday's flask cultivation and a sample of equal size of E. coli KRX fermentation with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] were disrupted via sonication and ECOL was purified by using the HisTrap column. The purificated sample of the fermentation did not show any activity, so we decided not to purify the whole sample.
- First fermentation of E. coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000]
- Settings: fermenter: Braun Biostat B, autoinduction medium, final volume: 3 L, 37 °C, stirrer on cascade to hold a pO2 of 50 %, airflow: 2 NL/m, 12 hours.
- A growth kinetic was determined by measuring the OD600 every 30 minutes.
Saturday September 1st
- Team Cloning of Bacterial Laccases:
- Plasmid isolations on the plated colonies and control restriction with NotI. Control digest showed for [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 J23100 + J61101+ tthl_HIS], J23100 + J61101 + ecol_HIS positive bands. So we sent these plasmids for sequencing.
- Team Fungal and Plant Laccases: Plating two colonies from tvel35+pSB1C3 transformation on LB +CM agar.
- Team Cellulose Binding Domain:
- Got green colonies on [http://partsregistry.org/Part:BBa_K863122 const.GFP_His]-dish. Picked three and plated them on CM-select-agar.
- Team Cultivation & Purification:
- Harvesting and centrifugation culture of fermentation on 08/31 (E.coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000]). Store pellet at 4 °C.
- Team Substrate Analytic:
- Dominik instructed us, that we could operate the HPLC on our own. We also learnd how to clean the HPLC vials.
Sunday September 2nd
- Team Cloning of Bacterial Laccases:
- Control digest of tvel35 in pSB1C3 was positive.
- Plasmid isolation from pSB1C3 + J23110, pSB1C3 + J23103 and J23100 + J61101+ bpul_HIS. Control digest showed correct bands for J23100 + J61101+ bpul_HIS, pSB1C3 + J23103 and pSB1C3 + J23110.
- Team Cellulose Binding Domain:
- unsuccessful Colony-PCR of [http://partsregistry.org/Part:BBa_K863113 CBDclos(T7)+GFP_His].
- Team Site Directed Mutagenesis:
- Gradient-PCR of tvel10-plasmid with tvel10-t243g primer-mix.
- Team Cultivation & Purification:
- Made precultures of E. coli KRX containing either [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] or [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863121 BBa_K863121].
- Team Activity Tests:
- This week we had Team Modeling over and they told us about their concerns. To continue modeling they wanted to have a look at the activity of our laccase from T. versicolor but with different ABTS concentrations. Especially the were interested in the first time points after adding ABTS. This should give them enough information to calculate the enzyme activity. We didn't want to wait, so we started immediately with our standard activity test. Our tested ABTS concentrations were: 0.5µl, 1µl, 2µl, 4µl and 8µl. We got nice activity curves but also noticed, that the activity saturated quickly and therefore the initial activity of our laccase can not be measured accurately. Of course Team Modeling got our data just in time, but we also want to start new activity tests with half of the amount of laccase. So we are still trying to keep our lovely Team Modeling satisfied.
Summary of Week 19
hier eine Zusammenfassung
Week 19 (09/03 - 09/09/12)
Contents |
Weekly Seminar
- We added a band for our sponsors on every wiki-page.
- On thursday we will meet with René Röspel, member of the german parliament.
- We are invited to the annual reception of the University Bielefeld on the 5th of october. Well that won't work for us.
- We still are working on a design for our team t-shirt.
Monday September 3rd
- Team Cloning of Bacterial Laccases:
- We sent different plasmids for sequencing.
- We got another E.coli strain (Rosetta-gami 2) from our supervisor Dr. Christian Rückert. The cells carry a chloramphenicol-resistant plasmid, pRARE2, which supplies tRNAs for seven rare codons, AUA, AGG, AGA, CUA, CCC, GGA and CGG under the control of their native promoter. We want to use this strain for the expression of TTHL and BHAL hopeful that some rare codons are the reason that the laccases can’t be expressed by now. So we need the genes under control of a constitutive promoter because this strain has no T7 polymerase and we can’t use our inducible constructs. Further the plasmids need to have AMP resistance, because otherwise we can't select on positive transformed cells. Hopefully the plasmids, we sent for sequencing are ok, so we can use them.
- Team Cellulose Binding Domain:
- A autoinduced culture of KRX with the [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His]-plasmid does not seem to produce the protein (no green fluorescence).
- Team Site Directed Mutagenesis:
- Clean-up of tvel10-t243g-PCR
- pfu-gradient-PCR of xccl-plasmid with xccl-primers with no product.
- Team Cultivation & Purification:
- Made one half of the SDS-PAGES of the flask cultivation from 08/30.
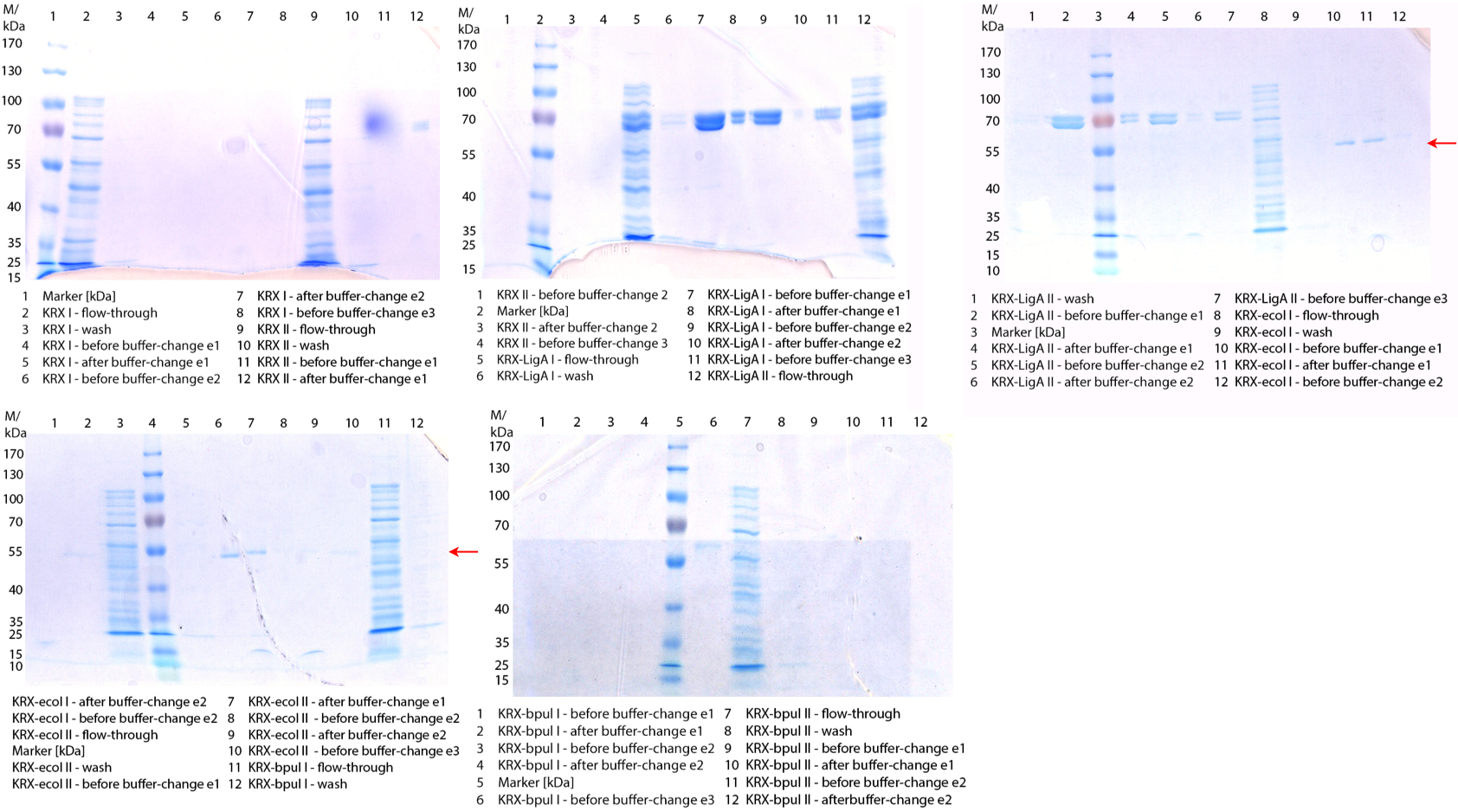 Figure 1: SDS-PAGES of purification from cultivation (08/28, smaller scale but with a double determination) of E.coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] or [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020] with positive ([http://partsregistry.org/Part:BBa_K525710 BBa_K525710]) and negative control (E.coli KRX without plasmid). 60 mL in 300 mL flasks without baffles with autoinduction medium, 60 µg mL-1 chloramphenicol at 37 °C for 12 hours. Purification of the supernatant via syringe method. The arrow shows ECOL (53 kDa)
Figure 1: SDS-PAGES of purification from cultivation (08/28, smaller scale but with a double determination) of E.coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] or [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020] with positive ([http://partsregistry.org/Part:BBa_K525710 BBa_K525710]) and negative control (E.coli KRX without plasmid). 60 mL in 300 mL flasks without baffles with autoinduction medium, 60 µg mL-1 chloramphenicol at 37 °C for 12 hours. Purification of the supernatant via syringe method. The arrow shows ECOL (53 kDa) - Work on culture of E. coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] (08/31) was continued and we performed the cell disruption by high-pressure homogeniser in 3 cycles. Then it was purificated by using a Ni NTA column. The product was given to the activity team for analysis.
- Fermentation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863121 BBa_K863121] for the cellulose binding team.
- Settings: fermenter: Braun, autoinduction medium, 60 µg/mL chloramphenicol, 3 L, 37 °C, stirrer on cascade to hold a pO2 of 50 %, airflow: 2 NL/m. This time we cultivated only for 9 hours and then decreased the temperature at 20 °C for a better folding of the protein for 1 hour. We had some problems with cascade at the end of the fermentation.
- Cells of today's fermentation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863121 BBa_K863121] were disrupted via high-pressure homogeniser and purified by using the Ni NTA column. The flow-through fluoresced very strong, so that not that much of the protein had bound to the column. Conclusion: The column had to be reloaded.
- Fermentation of E. coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000].
- Settings: fermenter: Braun, autoinduction medium, 60 µg/mL chloramphenicol, 3 L, 37 °C, stirrer on cascade to hold a pO2 of 50 %, airflow: 2 NL/m, 9 hours. We only cultivated 9 hours, because we saw that a great amount of our cells already died.
- Made precultures of E. coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] and one containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863113 BBa_K863113].
- Made one half of the SDS-PAGES of the flask cultivation from 08/30.
- Team Activity Tests:
- Today our hard-working Team Cultivation offered us some [http://partsregistry.org/wiki/index.php/Part:BBa_K863000/ BPUL] laccases. After fermentation and Ni NTA purification the laccase was still hanging around in imidazole buffer, so we rebuffered it into nanopure water. We incubated the sample with copper and considered to measure the protein concentration before running the activity tests. Using the protocol for determining the protein concentration via Bradford (standards were 1 mg/ml, 1.5 mg/ml and 2 mg/ml BSA) we got a final [http://partsregistry.org/wiki/index.php/Part:BBa_K863000/ BPUL] laccase concentration of 0.22 mg/ml. With this information we designed the following activity test different as usual. As a positive control we took 140 µl of our bought laccase TVEL0 with a concentration of 0,021 mg/ml. Accordingly to this concentration we applied 14 µl of the [http://partsregistry.org/wiki/index.php/Part:BBa_K863000/ BPUL] laccase to get approximately the same amount of laccase into each well. Additionally we tested 140 µl of the [http://partsregistry.org/wiki/index.php/Part:BBa_K863000/ BPUL] laccase to make sure that we will definitely see any activity. After measuring the samples the whole night we got our results in the morning. It turned out, that our store bought laccase TVEL0 was rapidly active as seen before in our activity tests. The [http://partsregistry.org/wiki/index.php/Part:BBa_K863000/ BPUL] laccase was also active. Using 140 µl of it led to the maximal state of oxidation of ABTS after 45 minutes. With the reduced amount of 14 µl of laccase this optimum was reached after 90 minutes. For our following activity tests we want to include the protein concentration again.
Tuesday September 4th
- We signed in for our tracks today! Our first choice is the 'Environment' track since it is obvious that we want to clean up drinking water and therefore help our environment. The second choice is the 'New Application' track. We are hoping our filter system can be applied in wastewater treatment plants and establishes a new area of water purification.
- Team Shuttle Vector:
- PCR Clean up was done for tAOX1 Part [http://partsregistry.org/Part:BBa_K863203 K863203].
- PCR of the fragment of tAOX1 for the shuttle vector was done with addition of betain (10 µL) and DMSO (1 µL) refered to 25 µL sample volume.
- Team Cloning of Bacterial Laccases:
- Digest of pSB1C3 + J23110 and pSB1C3+ J23103 for suffix insertion of the laccase genes.
- Digest of the different laccase ORFs for cloning in pSB1C3 backbone without any regulatory elements. Ligation of tthl, bhal, ecol and bpul in pSB1C3.
- Transformation of ligations from day before (J23100+J61101+pSB1A2+bhal_His).
- Team Site Directed Mutagenesis:
- plated six colonies of tvel-t243g for plasmid-isolation.
- Team Cultivation & Purification:
- Second half of the SDS-PAGES of the flask cultivation from 08/30 was made.
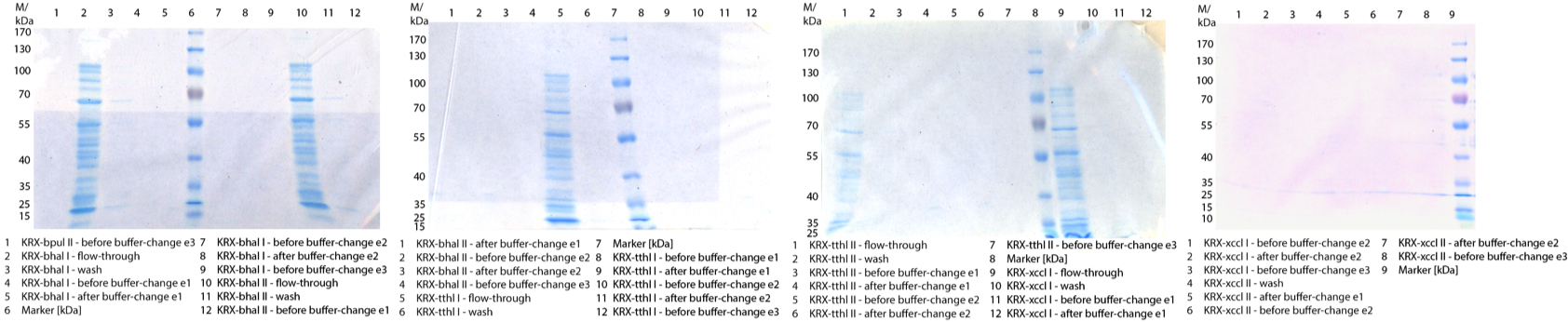 Figure 3: SDS-PAGES of purification from cultivation (08/28, smaller scale but with a double determination) of E.coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] or [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020] with positive ([http://partsregistry.org/Part:BBa_K525710 BBa_K525710]) and negative control (E.coli KRX without plasmid). 60 mL in 300 mL flasks without baffles with autoinduction medium, 60 µg mL-1 chloramphenicol at 37 °C for 12 hours. Purification of the supernatant via syringe method.
Figure 3: SDS-PAGES of purification from cultivation (08/28, smaller scale but with a double determination) of E.coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863010 BBa_K863010], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863015 BBa_K863015] or [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863020 BBa_K863020] with positive ([http://partsregistry.org/Part:BBa_K525710 BBa_K525710]) and negative control (E.coli KRX without plasmid). 60 mL in 300 mL flasks without baffles with autoinduction medium, 60 µg mL-1 chloramphenicol at 37 °C for 12 hours. Purification of the supernatant via syringe method. - After reloading we repeated the purification of BPUL produced by fermentation on 08/31. Therefore the flowthrough and the washing fraction were used. The different fractions should be tested by the activity team.
- A new fermentation of E. coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] was started
- Settings: fermenter: Infors, autoinduction medium, 60 µg/mL chloramphenicol, 3 L, 37 °C, stirrer on cascade to hold a pO2 of 50 %, airflow: 2 NL/m, 9 hours. We shorten the time, because the effect should be similar to the production of BPUL.
- The fermentation of E. coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] and the one containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] were harvested and after centrifugation stored at 4 °C, because our high-pressure homogeniser is out of work.
- Made a flask cultivation for the cellulose binding team: cultivation of E. coli KRX containing the <partinfo>BBa_K863103</partinfo>-plasmid to express a cellulose binding domain (CBD) fused to a His-tagged GFP.
- Settings: 1 L flask with baffles, autoinduction medium, 60 µg/mL chloramphenicol, final volume: 250 mL, 37 °C, 120 rpm, 9 hours. We made a three-fold determination, but did not get fluorescing cultures. The problem is to purify this product, because it might be impossible to remove the protein from the column. If it binds via CBD and not via His-tag.
- Made precultures of E. coli KRX containing plasmids with laccases from ECOL from E. coli BL21 (DE3), B. pumilus DSM27 and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012] behind the constitutive promoter J23100 as well as one containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000]
- Second half of the SDS-PAGES of the flask cultivation from 08/30 was made.
- Team Substrate Analytic:
- Today we let TVEL0 degrade Estradiol and Estrone. We prepared seven 2 mL Eppendorf tubes with: 425 µL BR-buffer, 50 µL TVEL0 and 25 µL Estradiol. To stop the reaction we added 1,5 mL MeOH. The Estradiol concentration should be 2 µg ml -1. We stopped at houre 0, 1, 2, 3, 4, 5, and over night, and mesured the Estradiol concentration with the HPLC and the LCMS.
Wednesday September 5th
- Team Cloning of Bacterial Laccases:
- Transformation of tthl, bhal, ecol and bpul in pSB1C3.
- Team Cultivation & Purification:
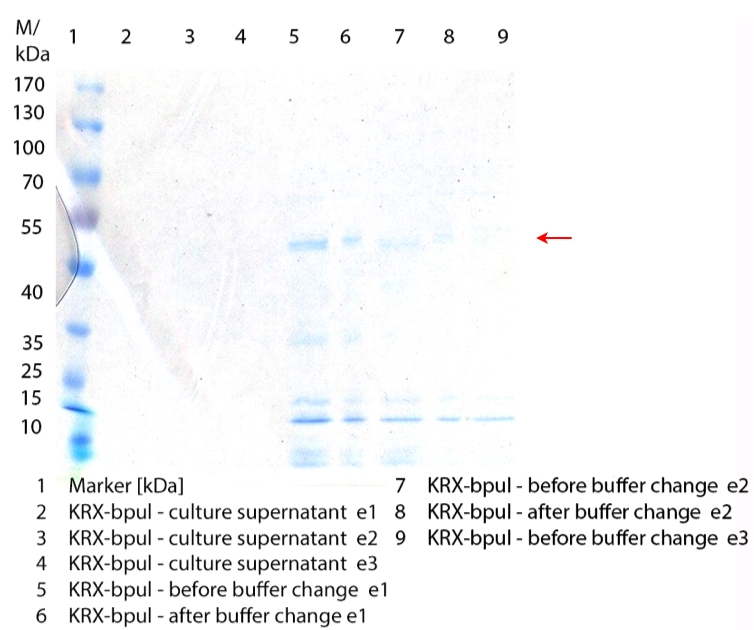 Figure 4: SDS-PAGE of purification from our first fermentation of E. coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] (08/31). 3 L fermenter (Infors), 37 °C, pO2 of 50 % for 12 hours. Purification of the supernatant via syringe method. The arrow shows BPUL (58.6 kDa) in lane 5-8 (elution 1 and 2).
Figure 4: SDS-PAGE of purification from our first fermentation of E. coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] (08/31). 3 L fermenter (Infors), 37 °C, pO2 of 50 % for 12 hours. Purification of the supernatant via syringe method. The arrow shows BPUL (58.6 kDa) in lane 5-8 (elution 1 and 2).- Performed the cell disruption of yesterday's flask cultivation to produce CBD-GFP+His fusion protein via sonication and made a SDS-Page of it. The problem is to purify this product, because it might be impossible to remove the protein from the column. If it binds via CBD and not via His-tag.
- Made a flask cultivation of E. coli KRX containing new BioBricks: Laccase ECOL from E. coli BL21 (DE3), BPUL from B. pumilusDSM27 and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012] behind the constitutive promoter J23100.
- Settings: 100 mL flask without baffles, autoinduction medium, 60 µg/mL chloramphenicol, final volume: 60 mL, 37 °C, 120 rpm, 12 hours, double determination.
- We repeated the purification of His-tagged GFP by using the flow-through and the washing fraction, but it did not work. Then we got to know that it could not work, because the sequencing showed us that the GFP was not tagged to His.
- Made a SDS-Page of the fractions of purified B. pumilus laccase produced at 08/31.
- Starting of another fermentation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000].
- Settings: fermenter: Braun, autoinduction medium, 60 µg/mL chloramphenicol, 3 L, 37 °C, stirrer on cascade to hold a pO2 of 50 %, airflow: 2 NL/m, 6 hours. Found out that we got a maximal optical density after 4 hours, after which the cells start to die. Therefore we harvested the cells after 6 hours.
- Aim: Getting a growth kinetics.
- Made preculture of E. coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005]
- Team Activity Tests:
- Today we had to deliver some data for Team Modeling. To calculate enzyme kinetics of our laccases the initial reaction speed has to be determined. With our usual approach using 0.1 U TVEL0 Laccase we couldn't accurately measure the first minute of reaction with ABTS because everything went to fast. So we cut down the volume of TVEL0 laccase from 140 µL to 35 µL (0.025 U) and measured the oxidizing potential of TVEL0 with different ABTS concentrations. The results are shown in Fig. 5. There you go, Team Modeling! Make the best out of it!
- Team Substrate Analytic:
- The degradations from the 4th September were stopped with methanol. The samples were evaporated for six hours and diluted in 50 % - 50 % acetonitril - water. Then we used the C18E-columns for purifying the samples. The measuring with LC-MS was done from Marcus Persicke.
- It´s time to regenerate the column. Dominik showed us the protokol.
Thursday September 6th
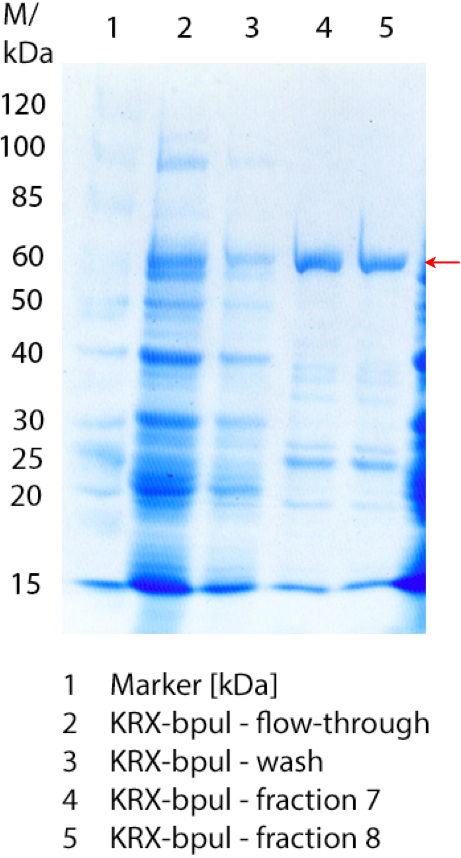
- Team Cloning of Bacterial Laccases:
- Colony PCR on [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863022 J23100 + J61101 + bhal_HIS] showed four positive colonies which were plated on LB with CM + AMP.
- Plating colonies from colony PCR on bpul+ pSB1C3 and tthl + pSB1C3 on new plates.
- We still have to wait for the sequencing results but control restrictions suggests that we have tthl_HIS, bhal_HIS, ecol_HIS and bpul_HIS in a vector with a constitutive promoter. So we transformed the plasmids in E.coli Rosetta Gami 2. The cells carry a chloramphenicol-resistant plasmid, pRARE2, which supplies tRNAs for seven rare codons, AUA, AGG, AGA, CUA, CCC, GGA and CGG under the control of their native promoter.
- Team Cellulose Binding Domain:
- gradient-PCRs to generate [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg] and [http://partsregistry.org/Part:BBa_K863111 CBDclos_Freiburg] on the orignial plasmids with the CBDcex_Freiburg + CBDclos_Freiburg and 2AS_Freiburg-Reverseprimers. Only the [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg]-product could be generated (68,5°C is best).
- two colonies of [http://partsregistry.org/Part:BBa_K863122 const.GFP_His] plated on selection-agar for plasmid-isolation.
- Team Cultivation & Purification:
- Made a SDS-PAGE of cultivation from 08/31.
- Fermentation of E. coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] to get a growth kinetics
- Settings: fermenter: Infors, autoinduction medium, 60 µg/mL chloramphenicol, 3 L, 37 °C, stirrer on cascade to hold a pO2 of 50 %, airflow: 2 NL/m.
- Preparation of the bigger fermenter (7 L) for the first time
- Performing the cell disruption via sonification and the purification via HisTrap column supported by Äkta of the flask cultivation from 09/05.
- We got to know that the working group of physical chemistry has another homogeniser, that we could use. We performed the cell disruption via high-pressure homogeniser of fermentations from 09/03, 04 and 05.
- Made preculture of E. coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005]
- Team Substrate Analytic:
- The fluorescent lamp is broken. So it was not possible to mesure today.
Friday September 7th
- Team Cloning of Bacterial Laccases:
- Plasmid isolation of different plated colonies and control digest. The restriction showed positive results for the plasmids for [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863001 bpul+pSB1C3], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863011 tthl + pSB1C3] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863022 bhal_His + J23110 + RBS + pSB1C3]. So we sent the plasmids for sequencing.
- Transformation of <partinfo>BBa_K863012</partinfo> and <partinfo>BBa_K863022</partinfo> in E. coli Rosetta-Gami 2.
- Team Shuttle Vector:
- Designed and ordered sequencing primers for the shuttle vector. We set a primer every 700 to 750 bases.
- Now all fragments are gel purified and the second Gibson assembly could be started. The fragments were mixed equimolar as described below with 15 µL of the Gibson master mix:
| Fragment | Quantity [ng] | Volume [µL] |
|---|---|---|
| 5AOX1 | 26.5 | 0.44 |
| MFalpha1 | 8.3 | 0.11 |
| 3AOX1 | 19.4 | 0.44 |
| HIS4 | 73.0 | 2.16 |
| pSB1C3 | 55.44 | 1.54 |
| tAOX1 | 8.9 | 0,11 |
After incubation for 1 h at 50 °C, 2 µL were transformed in E. coli KRX cells by electroporation. And plated on selective LB Cm plates.
- Team Cultivation & Purification:
- Today we performed lots of purifications. This time we used a Ni-NTA column, because it seemed to work better than the TALON column. Therefore it was reloaded newly before starting the purifications. The purified samples were BPUL produced on 09/03 and 09/05 as well as ECOL produced on 09/04. The results were not that promising, so maybe we harvested the cells too early. They start dying more or less when we expect them to produce our (toxic) protein. So we think we had to let some of them die, so that the rest will produce our protein. The next fermentations should endure at least 12 hours.
- Fermentation of E. coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] within the Bioengineering NFL22 fermenter.
- Settings: fermenter: Biostat NFL22 (7 L), autoinduction medium, 60 µg/mL chloramphenicol, final volume: 6 L, 37 °C, stirrer increased 2 % if the pO2 got below 30 %, airflow: 5 NL/m, 12 hours.
Saturday September 8th
- Team Shuttle vector:
- Colonies were picked and Colony PCR was done. Some positive clones could be identifed and were plates out for plasmid isolation.
- Team Cultivation & Purification:
- Harvesting and centrifugation of fermentation of 09/07. Pellet stored at 4 °C.
- Made precultures of E. coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] and of E. coli Rosetta-Gami 2 containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012] and laccase from B.halodurans <partinfo>BBa_K863022</partinfo>.
- Team Activity Tests: Today we managed to rebuffer the [http://partsregistry.org/wiki/index.php/Part:BBa_K863000/ BPUL] samples given by Team Cultivation. We got fraction 3 to 7 and 15 to 19 of the purification, because they have shown a nice peak. After bringing the [http://partsregistry.org/wiki/index.php/Part:BBa_K863000/ BPUL] laccases into water, we incubated with 0.5 mM CuCl2 and measured the protein concentration via Bradford. The offered fractions contained the following protein concentrations after rebuffering: 0.0087 mg/mL (fr. 3), 0.0110 fr. 4), 0.0632 (fr. 5), 0.1672 (fr. 6), 0.1229 (fr. 7), 0.0060 (fr. 15), 0.0034 (fr. 16), 0.0326 (fr. 18), 0.0225 (fr. 19). The protein amount in fraction 17 couldn't be measured but we didn't leave it behind. We adjusted the protein concentrations to the concentration of TVEL0, but did also measurements using 140 µL of each sample, because the protein concentrations do not only represent gained laccases. The results of our plain activity measurement of the samples are demonstrated in Fig. 7 and Fig. 8 (positive and negative control shown in Fig. 3):
Fractions 6 and 7 show the highest activity leading to the assumption, that faction 7 contains as much protein as fraction 6 and that this amount is almost represented through [http://partsregistry.org/wiki/index.php/Part:BBa_K863000/ BPUL] laccase.
Additionally we were interested in the impact on copper incubation. We have chosen fr. 18 and compared the activity of a copper incubated and a not copper incubated sample. With a incubation time of 2 hours with 0.5 mM CuCl2 we can double the activity of the [http://partsregistry.org/wiki/index.php/Part:BBa_K863000/ BPUL] laccase (see Fig. 10). Another interesting topic was the impact of imidazole buffer on the [http://partsregistry.org/wiki/index.php/Part:BBa_K863000/ BPUL] activity. For that we tested fr. 7 and fr. 18 in imidazole buffer and rebuffered in H2O. The results we got were unambiguously: [http://partsregistry.org/wiki/index.php/Part:BBa_K863000/ BPUL] in imidazole buffer didn't show much activity. Unfortunately we can't omit the rebuffering.
Sunday September 9th
- Team Shuttle Vector:
- Plasmid from the positive clones was isolated.
- Team Cultivation & Purification:
- Fermentation of E. coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] (fermenter: Infors) or containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] (fermenter: Braun) to get a growth kinetics.
- Settings: autoinduction medium, 60 µg/mL chloramphenicol, final volume: 3 L, 37 °C, stirrer on cascade to hold a pO2 of 50 %, airflow: 2 NL/m, 12 hours.
- Harvesting and centrifugation of today's 3 L fermentations.
- Fermentation of E. coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000]
- Settings: fermenter: Bioengineering NFL22 (7 L), autoinduction medium, final volume: 6 L, 37 °C, pH 7 stirrer increased 2 % if the pO2 got below 30 %, airflow: 5 NL/m, 12 hours.
- Flask cultivation of E. coli Rosetta-Gami 2 cells containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012] and <partinfo>BBa_K863022</partinfo>.
- Settings: flask without baffles, LB medium, 200 mL, 60 µg/mL chloramphenicol and 300 µg/mL ampicillin, 37 °C, 120 rpm. We cultivated for 48 hours and stored the culture then overnight at 4 °C.
- Fermentation of E. coli KRX containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] (fermenter: Infors) or containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] (fermenter: Braun) to get a growth kinetics.
Summary of Week 20
hier eine Zusammenfassung
Week 20 (09/10 - 09/16/12)
Contents |
Weekly Seminar
- We are discussing our lab results. Some experiments have to be repeated, but we are on a good way.
Monday September 10th
- Team Shuttle Vector:
- The PCR amplicons from the Parts: 3AOX1, tAOX1, 5AOX1, MFalpha1 and HIS4 were double digested with EcoRI and PstI in buffer O over night at 37 °C.
- Team Site Directed Mutagenesis:
- Digestion of the six tvel10-plasmids. Three were unmutated and other three hadn't lost the illegal SpeI-restriction-site, but their second fragment was of a smaller size.
- plated three additional tvel-t243g-colonies.
- Team Cellulose Binding Domain:
- Sequencing results arrived: One [http://partsregistry.org/Part:BBa_K863102 CBDcex(T7)] is completely right!
- Nanodropping plasmids of isolated [http://partsregistry.org/Part:BBa_K863113 CBDclos(T7)+GFP_His] showed that the cells did not have a plasmid at all (selection-agar did not work)
- Gradient-PCR with [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)] as template and [http://partsregistry.org/Part:BBa_K863111 CBDclos_Freiburg]-primers did work just fine (best temperature 61,7°C); Digestion with XbaI and AgeI.
- Gel-Clean-up of [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg] and [http://partsregistry.org/Part:BBa_K863121 GFP_His]
- Digestion of [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg] with XbaI+AgeI.
- Digestion of [http://partsregistry.org/Part:BBa_K863121 GFP_His] with NgoMIV and PstI.
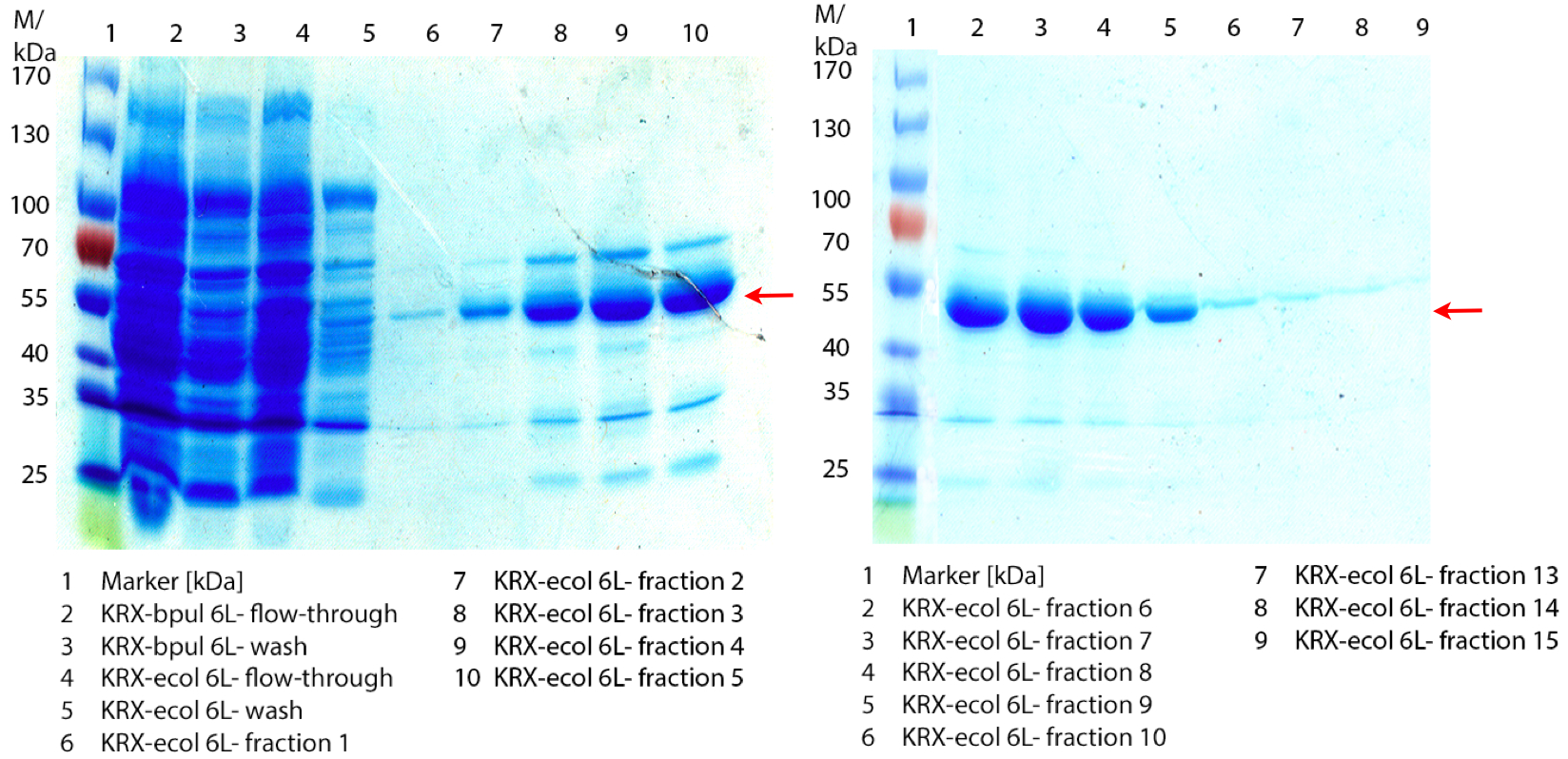
- Team Cultivation & Purification:
- Cell disruption via high-pressure homogenizer and purification via Ni-NTA column were performed for the following samples: 3 L fermentation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] (09/09), 6 L fermentation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] (09/07) and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] (09/09).
- We made SDS-PAGES of purification of ECOL.
Tuesday September 11th
- Team Shuttle Vector:
- Digest of shuttle shuttle vector with PvuII and HindIII as control. Agarose gel looks good.
- The parts (HIS4, 3AOX1, tAOX1, 5AOX1, MFalpha1) were ligated in pSB1C3 and transformed in E. coli KRX cells.
- Team Fungal and Plant Laccases:
- PCR on tvel5 laccase for cloning in shuttle vector. Digest of shuttle vector and digest of tvel35 with AarI enzyme, ligation and transformation in E. coli KRX cells.
- Team Cellulose Binding Domain:
- Assembly of <partinfo>BBa_K863104</partinfo> and <partinfo>BBa_K863114</partinfo>:
- Assembled [http://partsregistry.org/Part:BBa_K863111 CBDclos_Freiburg] and [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg] with [http://partsregistry.org/Part:BBa_K863121 GFP_His] and <partinfo>J61101</partinfo> and plated the ligation on AMP-selection-agar (because of the pSB1A2-backbone).
- Restriction of [http://partsregistry.org/Part:BBa_K863111 CBDclos_Freiburg] and [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg] with EcoRI and PstI.
- Ligation of CBDcex_Freiburg and CBDclos_Freiburg with the pSB1C3-backbone and transformated and plated on selection-agar
- Assembly of <partinfo>BBa_K863104</partinfo> and <partinfo>BBa_K863114</partinfo>:
- Team Cultivation & Purification:
- Made precultures of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 BBa_K863005] as well as of E. coli KRX.
Wednesday September 12th
- Team Fungal and Plant Laccases:
- Ligation of shuttle vector with tvel5 (digested with AarI).
- Team Site Directed Mutagenesis:
- Plasmid-isolation of the three tvel10-plasmids and digestion with SpeI showed two unmutated plasmids and one with the same wrong restriction-fragments as Monday. There must be a systematical error. pfu-PCR should be done again.
- Team Cellulose Binding Domain:
- There are only few colonies on all selection-agar-dishes, but none is obviously green fluorescing, even with UV-light emission could not be stimulated.
- Plated colonies of [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg] and [http://partsregistry.org/Part:BBa_K863111 CBDclos_Freiburg] to see if they are red or not.
- Colony-PCR of [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg] and [http://partsregistry.org/Part:BBa_K863111 CBDclos_Freiburg] showed eight positive colonies to the <partinfo>K863104</partinfo>-insert. Plated the positive colonies for plasmid-isolation.
- Team Cultivation & Purification
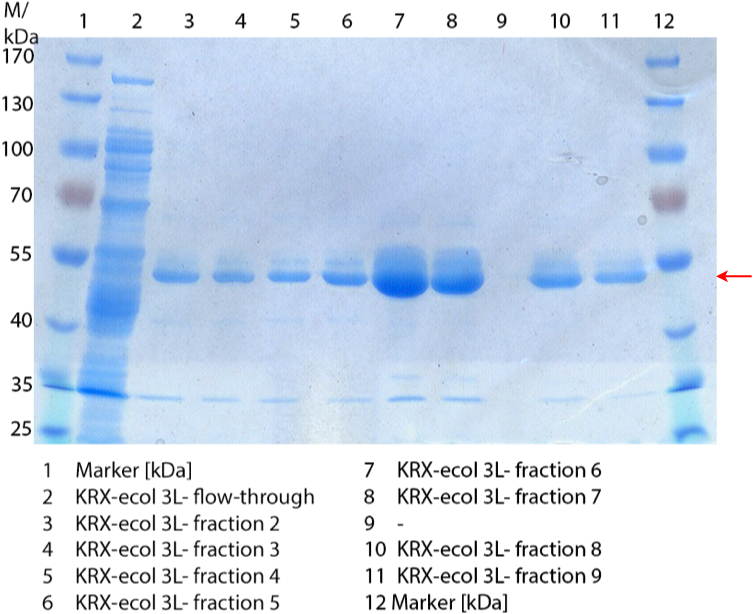
- Cell disruption via sonication and purification via Ni-NTA column were performed for the following samples: 200 mL cultivation of E. coli Rosetta Gami 2 containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012](09/09) and <partinfo>BBa_K863022</partinfo> (09/09) behind a constitutive promoter.
- Made SDS-PAGE of the fermentation from 09/09.
Thursday September 13th
- Team Fungal Laccases and Team Shuttle Vector:
- Bad day for us. We realized that our primers we used for amplification of the laccase genes for cloning in the shuttle vector don't have overhanging ends which were needed for an optimal activity of the enzyme AarI. So we had to order new primers with some bases prior to the recognition sites. But after there is little time left until wiki freeze and the new primers need some time until they are here we started a last trial for cloning this 'wrong' PCR products in the shuttle vector. The idea was to phosphorylate and ligate the ends of the PCR product from tvel5 together. Then the problem with the missing bases at the end would be solved and AarI can digest the end of the PCR product. We digested the shuttle vector with AarI and dephosphorylated it. After ligation we transformed the approaches in E. coli KRX cells.
- The ligation of the parts in pSB1C3 was not successful. So the Parts and the plasmid have to be digested with EcoRI and SpeI restriction enzymes.
- Team Cellulose Binding Domain:
- Designed a lot of primers to cope with the expression problem. E.g. inserting a long S3N10-Linker via blunt-end-cloning between the CBD and the GFP, also primers to get rid of the His-tag on the GFP and a last set to easily change the order of CBD and GFP (via assembly but with no linker in between).
- Plasmid-isolation of <partinfo>BBa_K863104</partinfo>-transformation clones
- Colony-PCR of [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg] and [http://partsregistry.org/Part:BBa_K863111 CBDclos_Freiburg] colonies. All [http://partsregistry.org/Part:BBa_K863111 CBDclos_Freiburg]-colonies are positive and half of the [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg]-colonies.
- Colony-PCR of CBDclos_F.+GFP with no positive result
- Colony-PCR of [http://partsregistry.org/Part:BBa_K863122 const.GFP_His]: 2 positive (one fluorescend); Plated both positive and one additional fluorescend.
- Team Cultivation & Purification:
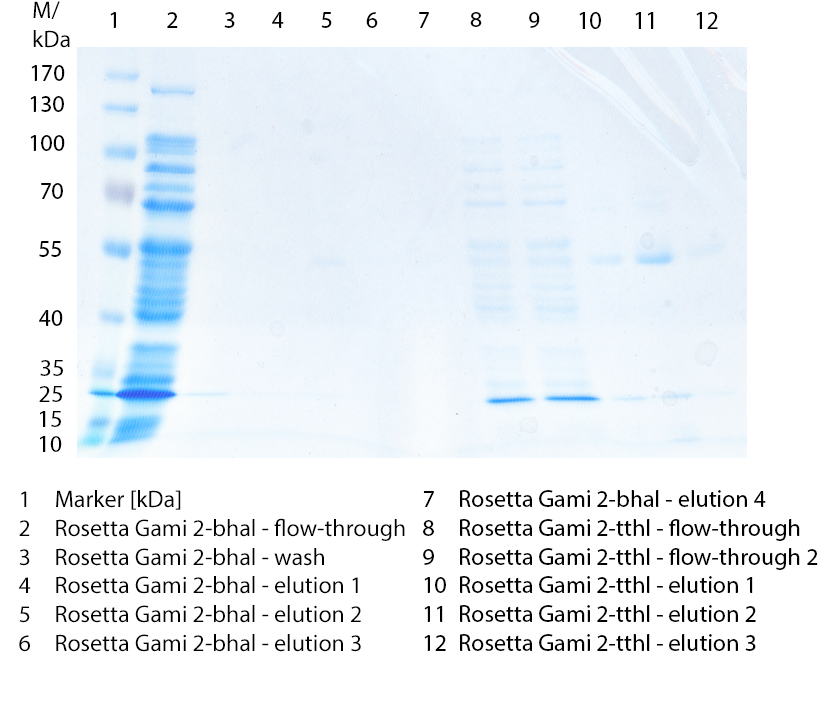
- SDS-Pages of the flask cultivation from 09/09 ( E. coli Rosetta Gami 2 containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012](09/09) and <partinfo>BBa_K863022</partinfo>)
- Fermentation of E. coli KRX without plasmid (fermenter: Infors) and with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000] (fermenter: Braun Biostat)
- Settings: fermenter: Infors/Braun Biostat, final volume: 3 L, autoinduction medium, 60 µg/mL chloramphenicol added, 37 °C, stirrer on cascade to hold a pO2 of 50 %, airflow: 2 NL/m, durance: 12 h.
- Made preculture of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863103 BBa_K863103] (CBD-GFP-His).
- Made preculture of P. pastoris GS115
- Fermentation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BBa_K863000].
- Settings: fermenter: Bioengineering NFL22(7 L), final volume: 6 L, autoinduction medium with 60 µg/mL chloramphenicol added, 37 °C, stirrer increased 2 % if the pO2 got below 30 %, airflow: 5 NL/m, 12 hours.
- Team Substrate Analytic: We did an Anthracen and Ethinylestradiol degradation to analyze them in LC-MS.
Friday September 14th
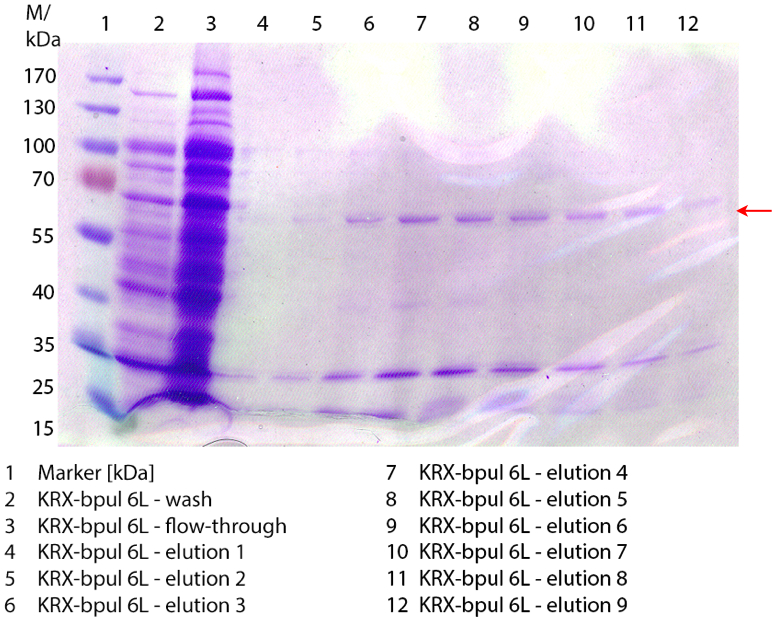
- Team Fungal Laccases and Team Shuttle Vector:
- Waiting for colonies from our transformation the day before.
- Ligate the new restricted parts in pSB1C3 and transformed them in KRX.
- Team Cellulose Binding Domain:
- Isolated three glowing dishes of KRX with the [http://partsregistry.org/Part:BBa_K863122 const.GFP_His]-plasmid, three with the [http://partsregistry.org/Part:BBa_K863111 CBDclos_Freiburg]-plasmid and [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg]-plasmid.
- Team Cultivation & Purification:
- Cell disruption of fermentation 09/13 via high-pressure homogenizer and purification via Ni-NTA column. Made SDS-Pages of purificated fractions.
- Repeat the preculture of P. pastoris GS115, because of using the wrong media.
- Made preculure of E. coli Rosetta Gami 2 with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012].
- Team Substrate Analytic:
We analyze Anthracen and Ethinyl estradiol from the 13th September. As you can see in the following figures Anthracen distingrates and Ethinyl estradiol on the other hand was stable. The corelation between the first peak and the other two peaks on Ethinyl estradiol may a pipetting mistake since the other two peaks are nearly simular. Distingration on Anthracen, a polycyclic aromatic hydrocarbon, were tested in different media and we determine that it depends on the Britton-Puffer (read more)
Saturday September 15th
- Team Cloning of Bacterial Laccases:
- Digest of <partinfo>BBa_J04450</partinfo> to have pSB1C3 backbone for the last clonings.
- Team Fungal Laccases and Team Shuttle Vector:
- We had some colonies on our LB + CM plates from our transformation of shuttle vector + tvel5. So we did Colony PCR to see if the gene was inserted. But sadly all colony PCRs are negative.
- Team Cellulose Binding Domain:
- KRX culture of [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)-GFP_His] seems to have a green glow.
- Isolation of four pellets of [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)-GFP_His] for us and SDU Denmark.
- [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)] digested with SpeI and AgeI and deposphorylated.
- [http://partsregistry.org/Part:BBa_K863121 GFP_His]-PCR-product (gel-clean) digested with SpeI and NgoMIV.
- Ligated [http://partsregistry.org/Part:BBa_K863112 CBDclos(T7)] with [http://partsregistry.org/Part:BBa_K863121 GFP_His] and plated on select-Agar.
- Restriction-analysis showed that all [http://partsregistry.org/Part:BBa_K863122 const.GFP_His]-plasmids are correct, as are the three [http://partsregistry.org/Part:BBa_K863111 CBDclos] and two of the [http://partsregistry.org/Part:BBa_K863101 CBDcex]
- Collected data to make a protocol for a Cellulose binding assay:
- Avicel: about 11,4 mg protein (CBD) binds to 1 g Avicel (0,14 mg/12,3 mg)
- Duration of incubation for CBD to bind to Avicel: about 30 minutes
- Washing and Lysis-buffer: 50mM Tris-HCl (pH 8.0)
- If needed: Elution with 80% ethylen-glycol (EG) or 1/5 Pellet to 4/5 EG (100%) of the overall volume.
- KRX culture of [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)-GFP_His] seems to have a green glow.
- Team Cultivation & Purification:
- Made competent P. pastoris GS115 cells.
- Fermentation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012]
- Settings: fermenter: Bioengineering NFL22 (7 L), final volume: 6 L, LB-medium with 60 µg/mL chloramphenicol and 300 µg/mL ampicillin added, 37 °C, stirrer increased 2 % if the pO2 got below 30 %, airflow: 5 NL/m. Problem: stirrer cascade did not work at the beginning.
- Harvest and centrifugation of cultivation & fermentations 09/14. Store pellet at 4 °C.
- Team Activity Test:
- Today we got the chance to work with Team Immobilization again. The samples they gave us were supernatants from different immobilization approaches. The team tested diverse bead concentrations with even higher amounts of beads then before. Check their labjournal for further information about the output of the measurements. Besides the supernatants we also received beads with immobilized laccases TEVL0 and [http://partsregistry.org/Part:BBa_K863005 ECOL]. So finally we got to use the filter plates that helped us to carry out the activity measurements and then stop the reaction at a precice point of time via centrifugation. Team Immobilization gave us enough samples for 7 measurements after different reactions times. We chose to stop the reaction after 2, 4, 8, 16, 32 and 64 minutes, respectively, to check the behavior of the immobilized laccase in relation to the time. Check their lab journal for the very exciting results!
Sunday September 16th
- Team Cellulose Binding Domain:
- Colony-PCR of 15 colonies from the [http://partsregistry.org/Part:BBa_K863113 CBDclos(T7)+GFP_His] transformation-plate (biotaq - Armin Nestat's recipe) with the result of a lot of positive clones.
- Prepared the sequencing of three [http://partsregistry.org/Part:BBa_K863122 const.GFP_His], three [http://partsregistry.org/Part:BBa_K863111 CBDclos_Freiburg] and [http://partsregistry.org/Part:BBa_K863101 CBDcex_Freiburg]
- Lysis of [http://partsregistry.org/Part:BBa_K863113 CBDcex(T7)+GFP_His] isn't glowing anymore, or never was.
- Team Cultivation & Purification:
- Harvest and centrifugation of fermentation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012]. Store pellet at 4 °C.
- Cell disruption via sonication and purification of cultivation of E. coli KRX with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863103 BBa_K863103] via Ni-NTA column.
Summary of Week 21
hier eine Zusammenfassung
Week 21 (09/17 - 09/23/12)
Contents |
Weekly Seminar
- Everyone has to change his/her information on the team-site and we are trying to fill in the polls of the other iGEM teams.
- We are discussing our lab results, same procedure as every week.
Monday September 17th
- Team Cultivation & Purification:
- Cell disruption of fermentation of E. coli Rosetta-Gami 2 with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012] via high-pressure homogenization and purification via Ni-NTA column.
- Team Activity Tests:
- Another day full of measurements is over! Today we measured some more samples from Team Immobilization: check their labjournal. As well we went on with characterizing our laccases. [http://partsregistry.org/Part:BBa_K863005 ECOL] was analyzed regarding its behavior in different concentrations of MeOH and acetonitrile. The activity was measured with concentrations of 2-16 µL, respectively. Since ABTS is changing it´s emission spectrum when being in contact with methanol we also detected the OD530. As expected ECOL is happier when only little MeOH is in its environment. We did the same measurements with acetonitrile and got the same result: the fewer acetonitrile, the better for ECOL. Again, we will hand over these results to Team Substrate Analytics.
- Also another measurement session started today. Team Cultivation lend their purified laccases from [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 TTHL] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863022 BHAL] produced with constitutive promoters to us. SO we rebuffered them and checked the activity with our standard protocol. And the results were good! In both cases we could detect activity in each fraction that Team Cultivation gave us (see Fig. 1 and Fig. 2). We are happy to have more laccases, but we are not sure, if we can pay enough attention to characterize them. We are trying our best, but [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BPUL] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863005 ECOL] come first.
Tuesday September 18th
- Team Shuttle vector:
The shuttle vector was linearized with EcoRI and SpeI and was transformed in competened P. pastoris cells.
- Team Cellulose Binding Domain:
- Primer arrived:
- Gradient-PCR with the [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His]-plasmid and S3N10-CBDcex_RV and S3N10-GFP_FW primer-mix. the right product appeared only at 66,5°C; Lower: wrong product; Higher: no product
- Following PCR at 66,5°C(50µL) had the wrong product (main product at 2 kbp); could be because of a different template concentration or not exact temperature.
- Alternativly took the 20 µL-tube with right product and added 0,5 µL DpnI for over-night digestion.
- Gradient-PCR with [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His]-template and a CBDcex_Freiburg- + GFP_Freiburg_compl-primer-mix to amplify the whole fusion-protein and get rid of the His-tag.
- Gradient-PCR with [http://partsregistry.org/Part:BBa_K863113 CBDclos(T7)+GFP_His]-template and a CBDclos_Freiburg- + GFP_Freiburg_compl-primer-mix to amplify the whole fusion-protein and get rid of the His-tag.
- Team Activity Tests:
- Today we measured the activity of our [http://partsregistry.org/Part:BBa_K863005 ECOL] laccase in a CuCl gradient to check whether another concentration than the one we usually incubated the laccases with might be more optimal. We chose a gradient from 0,1 mM to 0,7 mM (with an increment of 0,1) and measured the incubated samples as usual in 100 mM sodium acetate buffer, 0,1 mM ABTS (ad 200 µL H2O), respectively. Fig. 1 shows that the chosen concentration do influence ECOL, 0,4 mM seems like the best choice for incubation before the activity measurments. Thus the spectrum of OD420 maxima is not immensely wide so that [http://partsregistry.org/Part:BBa_K863005 ECOL] seems to be satisfied with any CuCl concentration.
- Additionally we measured the activity of [http://partsregistry.org/wiki/index.php/Part:BBa_K863010 TTHL] without constitutive promoter at 25°C and in sodium acetate buffer (pH 5). TTHL finally also shows some activity, our favorite fractions of Team Cultivations output are number 1 and 2. Unfortunately TTHL couldn´t be indentified correctly via MALDI yet so that we will have to wait with further characterizing until we get the go through the MALDI results.
- And another big suprise was the activity test of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BPUL]. After Team cultivation fermented it in a 6 L fermentation and purified our lovely enzyme, we did the standardized activity measurement protocol overnight. And have a look at our great results in fig. 6! We clearly have a good activity of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BPUL] reaching its saturation after half an hour. A fast reaction like this is comparable to the store bought laccase TVEL0. We are happy and will rebuffer as much as we can from fraction 4 which seems to have most of the Protein. Also we are going to determine the protein concentration. This is going to be awesome!
- Team Substrate Anayltics:
- We analyze Ethin estradiol and estradiol degradation with the Spectrofluorophotometer. The measure showed that the degradation product are detectable but the results weren't clear. We decide to do this measurement again to have better results so a new degradation of the two substrates were set.
Wednesday September 19th
- Team Cellulose Binding Domain:
- Stopped over-night-DpnI-digestion of S3N10-clos-GFP_his PCR-product and cleaned up in gel. Ligated the PCR-product with itself via Blunt-end-ligation and transformed it into KRX. Plated on Select-Agar.
- Gradient-PCRs of the [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His]-plasmid and a [http://partsregistry.org/Part:BBa_K863113 CBDclos(T7)+GFP_His]-plasmid (unsequenced) with the CBDcex_Freiburg- and GFP_Freiburg-compl- and the CBDclos_Freiburg- and GFP_Freiburg_compl primers, respectively. The CBDcex-setup showed no correct product, but the CBDclos-setup had the right bands at temperatures from 57°C to 64°C; Merged the correct fractions and cleaned them up through the gel.
- Digested the cleaned-up product with SpeI and XbaI and DpnI.
- Ligated <partinfo>J61101</partinfo> and [http://partsregistry.org/Part:BBa_K863111 CBDclos_Freiburg] with [http://partsregistry.org/Part:BBa_K863120 GFP_Freiburg] and transformed it into KRX.
- Team Activity Test:
- What a day! The deadline is coming closer and we still had (!) a lot of measurements to do, so today was chosen to be a day full of important measurements. First we measured the activity of the TVEL0 and [http://partsregistry.org/wiki/index.php/Part:BBa_K863000 BPUL] in a CuCl gradient. As well we examined what our [http://partsregistry.org/wiki/index.php/Part:BBa_K863000 BPUL] laccase does when its supposed to work in different pHs. [http://partsregistry.org/wiki/index.php/Part:BBa_K863000 BPUL] was additionally characterized regarding its behavior in different concentrations of MeOH and acetonitrile. This output is of course most interesting for Team Substrate Analytics. Since we already compared the behaviour of TVEL0 in the environment of sodium acetate buffer (pH 5) and Britton-Robinson buffer (pH 5), respectively, we also compared [http://partsregistry.org/wiki/index.php/Part:BBa_K863000 BPUL] and [http://partsregistry.org/Part:BBa_K863005 ECOL] in those buffers. ECOL is not impressed by the two different buffer systems and thus not influenced in it´s activity.
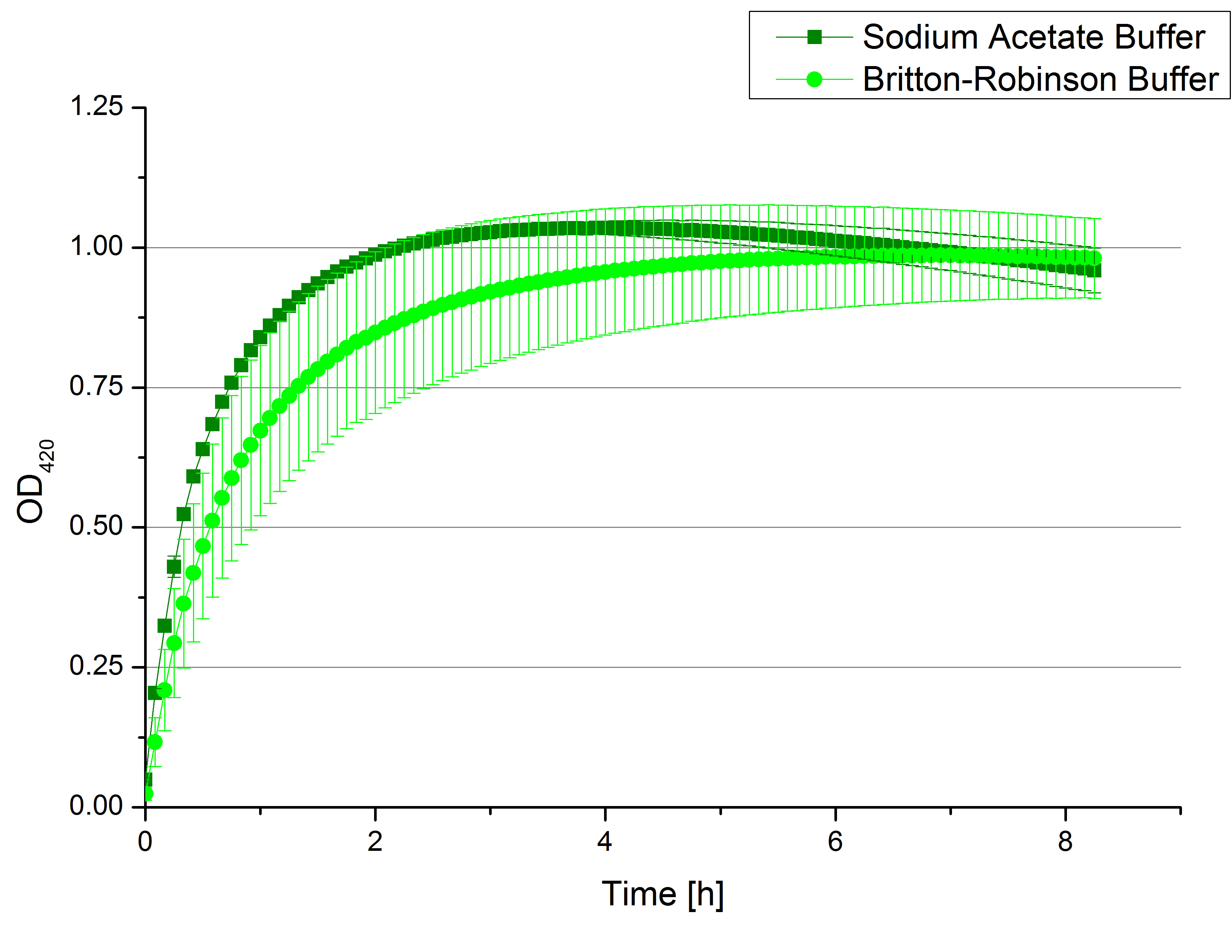
Thursday September 20th
- Team Fungal and Plant Laccases :
- Our primers for GFP, tvel35, tvel5 and <partinfo>BBa_K500002</partinfo> arrived. We did PCR on GFP with the new primers for cloning in our shuttle vector.
- Team Cellulose Binding Domain:
- No Colonies on the S3N10-[http://partsregistry.org/Part:BBa_K863113 CBDclos(T7)+GFP_His]-transformations-dish with the blunt-end ligation; reason: forgotten to phosphorylate the PCR-product.
- Two colonies on the CBDclos+GFP-dish with the constitutive [http://partsregistry.org/Part:BBa_J61101 J23100 + J61101] promoter. But none of them is glowing. And them shouldn't ... used the wrong selection-agar (since J61101 carries pSB1A2-backbone with AMP-resistence.
- Team Activity Tests:
- Today we started another exciting measurement. Today was all about temperatures. Through our cooperation with the clarification plant we found out that the waster water here in Germany has a minimal temperature of 10°C throughout the year. We used this value together with our previous measurements at 25°C to complete the temperature gradient of our measurements. Eventhough [http://partsregistry.org/Part:BBa_K863005 ECOL] shows an increased activity at 10°C, it does reach a comparable OD value as the samples measured at 25°C. The increased activity is expressed by achieving the maximal OD420 slower than the laccase measured at 25°C. This seems like good news for Team Modeling. On the other hand [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863000 BPUL] shows nearly the same great activity at 10°C as at 25°C. Of course the reaction is a bit slower, but nevertheless the saturation is reached after 3 hours.
- Team Substrate Analytics:
- Degradation measure from Ethinyl estradiol and Estradiol with the Spectrofluorophotometer. Degradation was from the 18th September
Friday September 21st
- Team Shuttle vector:
P. pastoris colonies are seen and are picked into liquid culture for genomic DNA isolation and genotype characterisation.
- Team Fungal and Plant Laccases:
- Digest of <partinfo>BBa_K863204</partinfo> and GFP PCR product with AarI and ligation. Transformation in E. coli KRX cells.
- Team Cellulose Binding Domain:
- Transformated <partinfo>J61101</partinfo> with CBDclos+GFP again and plated on the right selection-agar this time!
- Team Activity Tests:
- Another part of the characterization of our [http://partsregistry.org/Part:BBa_K863005 ECOL] and [http://partsregistry.org/wiki/index.php/Part:BBa_K863000 BPUL] laccase was to measure their activity when exposed to different concentrations of ABTS. We choose an exponential gradient of 2 µL to 16 µL and measured with the usual laccase concentration of 0,003 mg/mL and 100 mM sodium acetate buffer (ad 200 µL H2O). Unfortunately the measurements that contained 16 µL ABTS could not be measured completely because the OD420 exceeded the maximal value that Tecan was able to measure. The results of [http://partsregistry.org/Part:BBa_K863005 ECOL] (see. Fig. 1) show as well an exponential increase in the OD420 correlated to the ABTS concentrations, respectively. The negative control was chosen to contain the maximum of 16 µL ABTS in buffer and water that was incubated with CuCl2.
Saturday September 22nd
- Team Fungal and Plant Laccases:
- We did colony PCR on the transformed cells from day before. The results show that almost all colonies have the insert GFP. So we picked three of the positive colonies and plated each on five LB + CM plates, because ee need much vector for the transformation in Pichia pastoris.
- Team Cellulose Binding Domain:
- Once again two colonies on the agar-dish: can not say if they have a green glow...
- picked the colonies and them put into AMP-LB-Medium.
- Also made a flask with 10 mL LB-Medium and cells with the [http://partsregistry.org/Part:BBa_K863104 CBDcex(J61101)+GFP_His]-plasmid
- Once again two colonies on the agar-dish: can not say if they have a green glow...
- Team Cultivation & Purification:
- For collaboration with UCL London: Made precultures of E. coli KRX with <partinfo>BBa_K729006</partinfo>, with <partinfo>BBa_K863005</partinfo> and without plasmid.
Sunday September 23rd
- Team Cellulose Binding Domain:
- All cultures did not show any sign of GFP-glow under uv-light.
- Gradient-PCR of <partinfo>BBa_K863104</partinfo> with S3N10_Cex_compl- and S3N10_GFP-primermix to generate a 13 amino acid-linker between the cellulose binding domain and the GFP.
- Transformated [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His] and [http://partsregistry.org/Part:BBa_K863113 CBDclos(T7)+GFP_His] into BL21, to test if the problem is in the expression-system.
- Team Cultivation & Purification:
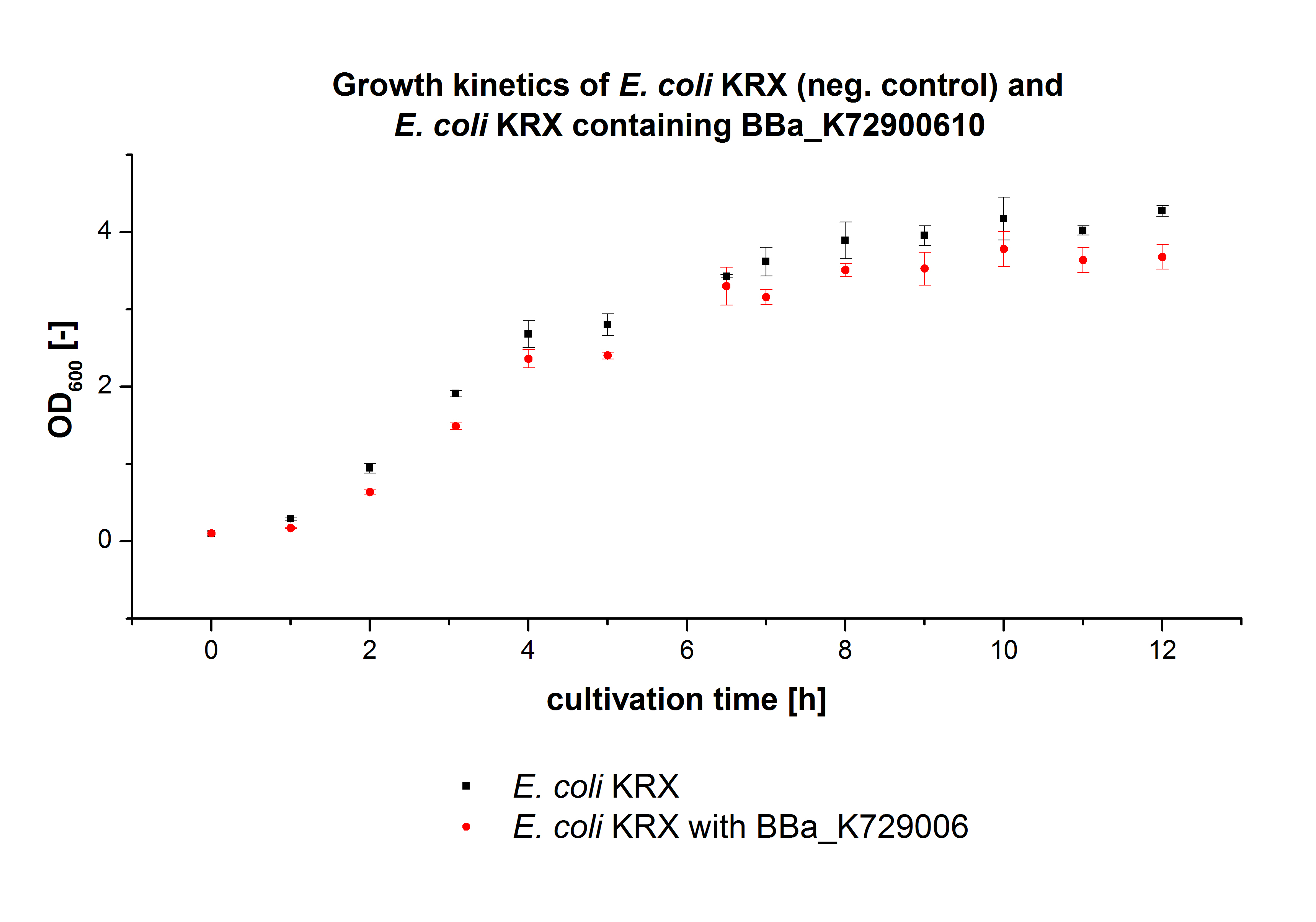
- For collaboration with UCL London: Cultivation of E. coli KRX with <partinfo>BBa_K729006</partinfo> and E. coli KRX without plasmid as negative control
- Settings: 250 mL flasks without baffles, final volume: 60 mL, LB-medium, 60 µg/mL chloramphenicol, 37 °C, durance: 12 hours.
- Settings: 1 L flask without baffles, final volume: 200 mL, LB-medium, 60 µg/mL chloramphenicol, 37 °C, durance: 12 hours.
- We determined the OD600 for a growth kinetics (see figure 1).
- Cultivation of E. coli KRX with <partinfo>BBa_K863005</partinfo> for comparison.
- Settings: 250 mL flasks without baffles, final volume: 60 mL, autoinduction medium, 60 µg/mL chloramphenicol, 37 °C, durance: 12 hours.
- Made SDS-Page of the cultivation from 09/15.
- For collaboration with UCL London: Cultivation of E. coli KRX with <partinfo>BBa_K729006</partinfo> and E. coli KRX without plasmid as negative control
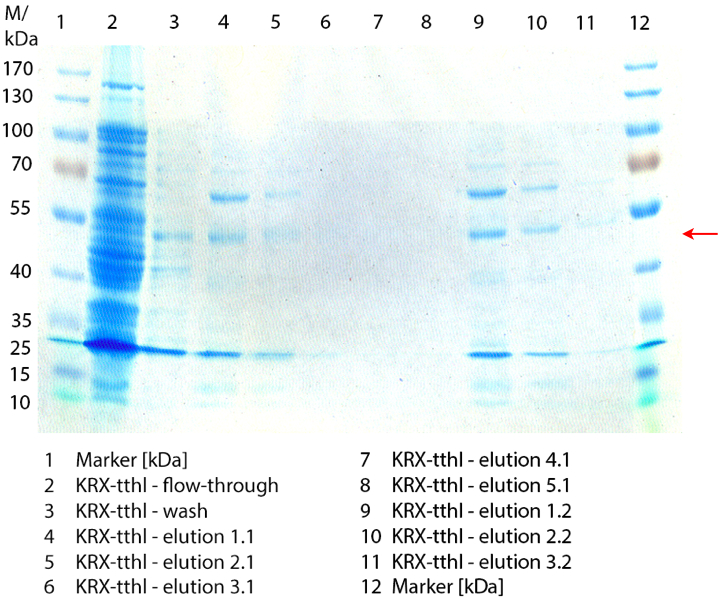
- Team Substrate Analytic: Since our last degradation with Estradiol didn't work because the Estradiol tubes were wrong we did an Estradiol degradation. We could find degradation products but not identify thoose products. Besides this we anayzed the sewage water we got from the Obere-Lutter and Schloß-Holte sewage water.
Summary of Week 22
hier eine Zusammenfassung
Week 22 (09/24 - 09/30/12)
Contents |
Weekly Seminar
- First plan for the team t-shirt: a pacman eating estrogen.
- We now gather every day 9 o'clock in the morning to work on the wiki.
- Julia V is implementing the last design changes to the wiki.
Monday September 24th
- Team Cellulose Binding Domain:
- There was no obvious glow in the IPTG induced cultures. Todays time is only enough to harvest. The cultures were centrifuged at top speed and the supernatant was discarded. Cell-pellets were stored at -20 °C for further measurements.
- Phosphorylation of the CDBcex-S3N10-GFP-PCR-product, followed by ligation, transformation in E. coli KRX and plating on select-agar.
- Team Cultivation & Purification:
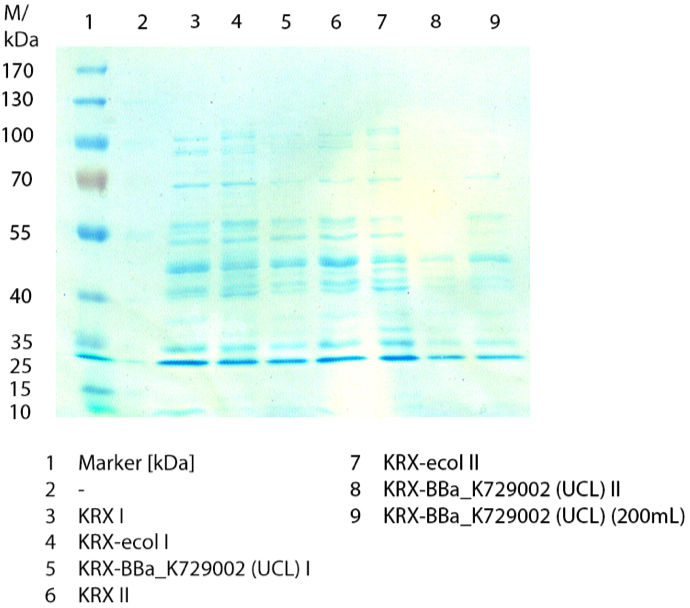 Figure 1: SDS-PAGE of the lysate supernatant from cultivation (09/23) of E. coli KRX with <partinfo>BBa_K729006</partinfo> , E. coli KRX without plasmid (negative control) and with <partinfo>BBa_K863005</partinfo> (positive control). 60 mL in 250 mL flasks without baffles,LB-medium or autoinduction medium for <partinfo>BBa_K863005</partinfo> , 60 µg/mL chloramphenicol, 37 °C for 12 hours.
Figure 1: SDS-PAGE of the lysate supernatant from cultivation (09/23) of E. coli KRX with <partinfo>BBa_K729006</partinfo> , E. coli KRX without plasmid (negative control) and with <partinfo>BBa_K863005</partinfo> (positive control). 60 mL in 250 mL flasks without baffles,LB-medium or autoinduction medium for <partinfo>BBa_K863005</partinfo> , 60 µg/mL chloramphenicol, 37 °C for 12 hours.- For collaboration with UCL London: Made celldisruption via sonication and SDS-Page from lysates. The bands were analysed by MALDI-TOF.
- Team Substrate Analytic: The sewage water has shown no substrates of our interests. We only had 1L of the sewage plant water, so it may depend on the low concentration that we couldn't detect anything. Estradiol degradation on the other hand has shown some peaks, probably from degradation products.
Tuesday September 25th
- Team Cellulose Binding Domain:
- No colonies on the select agar plate. Could have some reasons: Phosphroylation could be in-successful; expanding the time, or phosphorylating the primers before doing the PCR could be done. Also could be added to the mix since it has a [http://www.ncbi.nlm.nih.gov/pmc/articles/PMC210195/ positive effect on the phosporylation efficiency].
Wednesday September 26th
Thursday September 27th
Friday September 28st
Saturday September 29nd
Sunday September 30rd
| 55px | | | | | | | | | | |
 "
"