Team:UNAM Genomics Mexico/Project/Description
From 2012.igem.org

Project Description
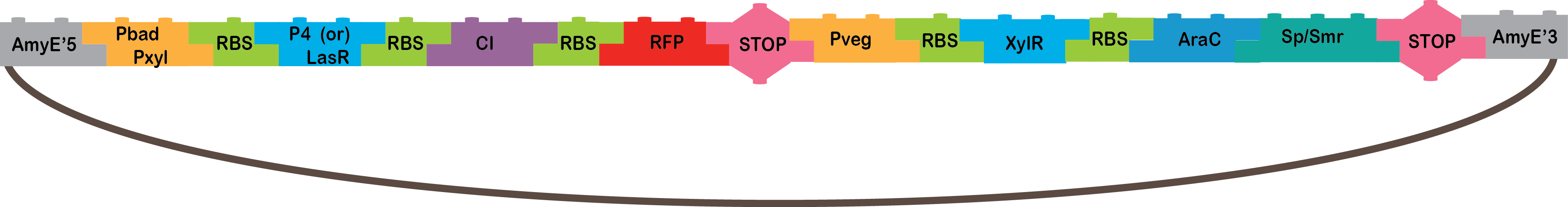
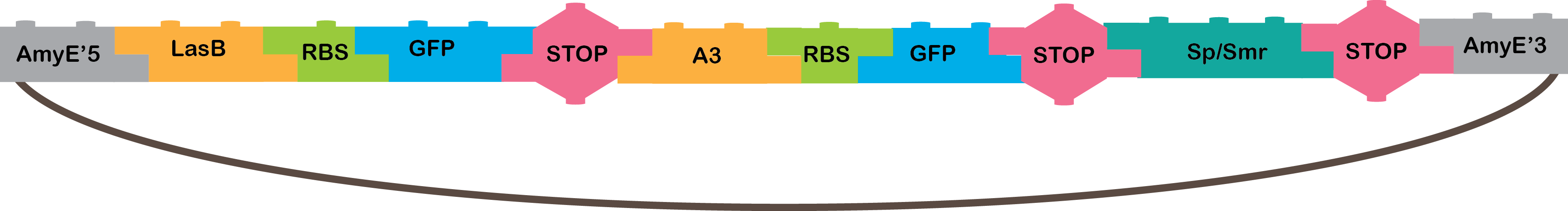
Our system consists of two different AND logic gates and an OR that derives from these ANDs. We expect three different Bacillus subtillis cells to be joined by nanotubes(1). Through these nanotubes, outputs from the two AND cells will pass to the cell containing the OR system and will function as inputs. The first cell will produce P4 after being induced by two different signals, it will not work with only one of these signals, hence the AND logic gate. The second cell will produce LasRin the same manner. Both P4 and LasR will pass through nanotubes to the third cell. This third cell functions as an OR logic gate. If either LasR or P4 pass through the nanotubes, they will activate promoters LasB and A3 respectively. These promoters will each induce the production of GFP, so if either LasR or P4 activate one of these promoters, we will prove that both and AND logic gate and an OR logic gate took place.
We designed two different AND systems which both produce LasR and P4. The difference between these two AND systems is how they are activated. The first AND has promoter pBad, which is induced by L-arabinose(2), and promoter pXyl which is induced by xylose (requiring transcriptional regulator XylR)(3). These two promoters are fused so that P4 or LasR will only be transcribed when both inductors are present. The second AND was inspired by Newcastle’s 2009 iGEM project(4). It also has a fused promoter, which consists of the ArsR promoter and CzrA. ArsR is induced by silver, arsenic, cadmium, or copper. CzrA is induced by zinc, cobalt, nickel, or cadmium5. Besides Newcastle’s 2009 design, we designed two different fused promoters with the same binding sites but in different order to try different combinations that could make the system more efficient. We wish to induce this system with two different metals to create the AND logic gate.
2Schlief, R. Regulation of the L-arabinose operon of Escherichia coli. Trends in Genetics, 2000; 16(12):559-565.
3[http://www.ncbi.nlm.nih.gov/pmc/articles/PMC210133/ Kreuzer P, Gärtner D, Allmansberger R, Hillen W. Identification and sequence analysis of the Bacillus subtilis W23 xylR gene and xyl operator. J Bacteriol. 1989 Jul;171(7):3840-5].
4[http://www.sciencedirect.com/science/article/pii/S0168952500021533 Newcastle University iGEM team. 2009. https://2009.igem.org/Team:Newcastle/Project#Cadmium_Sensing]
5[http://www.sciencedirect.com/science/article/pii/S1369527405000184 Moore CM, Helmann JD. Metal ion homeostasis in Bacillus subtilis. Curr Opin Microbiol. 2005 Apr;8(2):188-95].
 "
"