Team:METU/CellLimiter
From 2012.igem.org
Overview
In our project we aimed to construct a biofilm which will convert CO to CO2 biologically. Biofilms can be defined as the group of cells firmly united with the help of lipids, protein, DNA and polysaccharides and they can be used for beneficial applications [1] but the main obstacle in the case of biofilm formation is to keep the cell growth under control. We wanted our biofilm to work as a CO filter, due to that reason we designed a quorum sensing (QS) system to control the number of cells and to characterize the activity of biofilm better.
Cell population density changes as the bacteria grow and divide in the biofilm and QS can be defined as the regulation of gene expression during this change. As the cell density increases bacteria starts to release molecules named as autoinducers and according to the concentration of autoinducers gene expression is altered[2]. Eventually bacterial growth will be controlled. In our project we aimed to modify E.coli to synthesize its own QS signals, detect the cell density and prevent the cell division.
How The System Works ?
At the beginning of our QS system there is a constitutive promoter J23116 which is used for the constitutive production of LuxI, an autoinducer synthetase for AHL (acyl-homoserine lactone). Function of LuxI is to synthesize 3OC6HSL (3-oxohexanoyl-homoserine lactone). When 3OC6HSL is produced, it will form a complex with LuxR which is synthesized according to the activity of LuxPL promoter. After that point we aimed LuxR-3OC6HSL complex to increase the activity of LuxPR promoter. Finally LuxPR activity determines the amount of LasR and when PLasR promoter is activated MinC, a cell division inhibitor can be synthesized. Eventually, cell division stops after MinC reaches its critical concentration.
In our system antiholin is constitutively produced by the promoter J23116 and when antiholin level is in between 1000-3000 molecules, the cell will die.[4] Actually,it is the number of free holin molecules that determines if the cell will die or not.
Modelling
KILL-SWITCH MODEL
The aim of kill-switch model is to predict the behavior of kill-switch mechanism in different IPTG concentrations. We first tried to model mechanism by the help of ODEs. Then we used SimBiology toolbox of MATLAB to simulate system.Introduction:
As can be seen from the diagram our circuit involves constitutive production of LacI. LacI Promoter is negatively regulated by LacI. The control mechanism of our system is inhibition of LacI by IPTG molecule. Since LacI is inhibited by IPTG its effect on LacI promoter changes with respect to IPTG concentration.
Holin and Endolysin expression is controlled by cI promoter which is negatively regulated by cI.
Our first assumption in the model is that when the free Holin molecules, which means number of Holin molecules minus number of Antiholin molecules, reach 3000[ref] cell lysis occurs.
Because cI concentration depends on the activity of LacI promoter, we can conclude that in the presence of IPTG the concentration of cI will be high and expression of Holin and Endolysin will be lowered. Due to the constitutive production of Antiholin, we can say that free Holin molecules will be lowered and chance of cell lysis to occur will drop. However, to understand the overall behaviour of system with respect to IPTG concentration we need to model and simulate the system.

Mathematical Model:
In our model, we included all processes shown below.
Equations for mRNA transcription









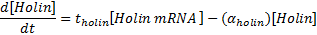


|
Parameter |
Description |
|
N |
Plasmid copy number |
|
Px |
Promoter Strength |
|
ax |
Degradation Rate |
|
Kx |
Disassociation (Equilibrium) Constant |
|
nx |
Hill Coefficient |
|
tx |
Translation Rate |
|
kx |
Binding on rate |
|
k-x |
Binding off rate |
|
Dx |
Diffusion rate |
Simulation Results
When the IPTG concentration is 0;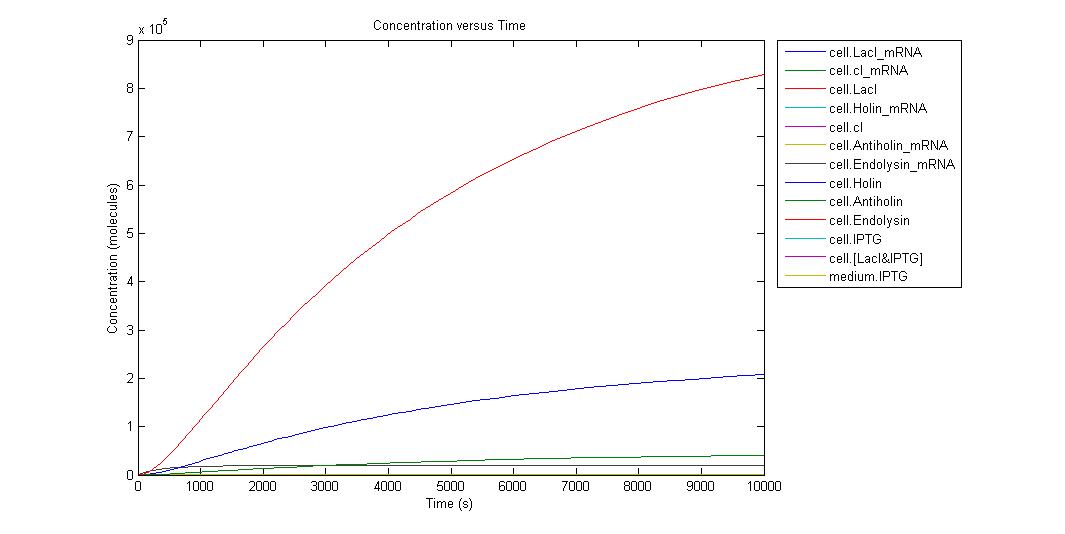
Individual graphs can be accessed here. Because the critical point in kill-switch is “Holin-Antiholin” molecules, we ran simulation in different IPTG concentrations and to understand result easier we select the outputs as Holin and Antiholin. Since translation rate of endolysin is more than Holin and Antiholin we can assume that there will be enough endolysin to make cell lysis occur. Also from the graph above we can understand that endolysin will be in sufficient amount.


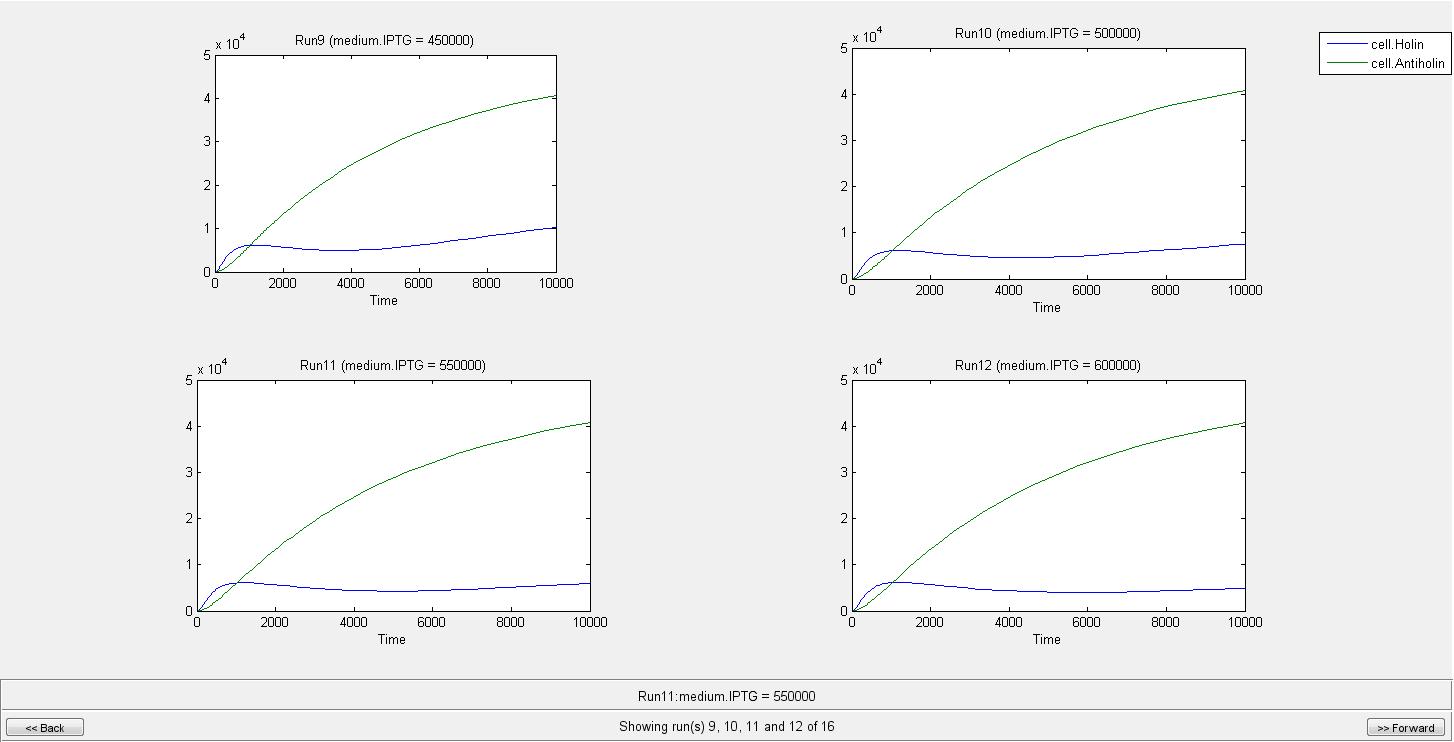

By the help of our model, we can conclude that when there is no IPTG molecule, cell lysis occur in 275 s. However when there is 100000 IPTG molecules, cell needs 2290 s to lyse. As can be seen from graphs until a very high IPTG concentration like in the Run14, the increase in Holin can eventually cause lysis. As a result of our model we concluded that after an IPTG concentration of 700000 molecule cell lysis will not occur. We are not considering the generation time of cell in this model since it can later be used for different organisms. If we consider e.coli with a generation time of 20 min, even in the medium with IPTG of 100000 molecules, it will divide before it can lyse.
When IPTG concentration is 700000;

Individual graphs can be accessed here.
Sensivity Analysis
Since the challenging part of modelling is choice of parameters, we conducted a sensitivity analysis to see how much result of our model depends on parameters. By the help of those, the critical values to be measured through experiments can be understood easily.






References
[1]Hansen LH, Knudsen S., Sorensen SJ. (1998). The effect of the lacY gene on the induction of IPTG inducible promoters, studied in Escherichia coli and Pseudomonas fluorescens. Current Microbiology,36 (6), 341-347. doi: 10.1007/s002849900320
[2]The fate of Gram-negative envelope during holin-endolysin lysis [Image]. (2000). Retrieved September 23, 2012, from: http://dx.doi.org/10.1016/S0966-842X(00)01705-4
[3]Tran TA, Struck DK, Young R. (2007). The T4 RI antiholin has an N-terminal signal anchor release domain that targets it for degradation by DegP. Journal Of Bacteriology, 189 (21), 7618-7625. doi: 10.1128/JB.00854-07.
[4]Christos G. Savva, Jill S. Dewey, John Deaton, Rebecca L. White, Douglas K. Struck, Andreas Holzenburg, Ry Young. (2008). The holin of bacteriophage lambda forms rings with large diameter. Molecular Microbiology, 69 (4), 784-793. doi: 10.1111/j.1365-2958.2008.06298.x
 "
"

