Team:University College London/LabBook/Week7
From 2012.igem.org



Contents |
Monday (23.7.12)
Aim - Picking colonies: On Friday 20.7.12 we found that the transformation for BBa_J23119, BBa_1750016, BBa_B0015, and BBa_B0034 all produced colonies and were suitable for colony picking.
(LOGO) Picking Colonies
Step 2 – Inoculating Colonies into a Selective Broth: The table below indicates the volume of broth and the concentration of antibiotic required for each BioBrick.
| Samples | Volume Inoculated | Broth (ml) | Antibiotic (ug/ml) | |
|---|---|---|---|---|
| BioBrick | BBa_J23119 | 10ul | Lysogeny Broth (5) | Ampicillin(50ug/ml) |
| 90ul | ||||
| BBa_I750016 | 10ul | |||
| 90ul | ||||
| BBa_B0015 | 10ul | |||
| 90ul | ||||
| BBa_B0034 | 10ul | |||
| 90ul | ||||
Tuesday 24.7.12
Aim – Results from Colony Picking
Results: The table below indicates whether there was growth for the BioBricks
| Samples | Volume Inoculated | Colony Formation | |
|---|---|---|---|
| BioBrick | BBa_J23119 | 10ul | Yes |
| 90ul | Yes | ||
| BBa_I750016 | 10ul | No | |
| 90ul | No | ||
| BBa_B0015 | 10ul | Yes | |
| 90ul | Yes | ||
| BBa_B0034 | 10ul | No | |
| 90ul | No | ||
Conclusion: We concluded that the failure of BBa_I750016 and BBa_B0034 is not necessarily due to failed transformation, as there was an unusual degree difficulty Picking the colonies 23.7.12. From this, it is possible that colonies were not properly inoculated, and so this requires a repeat. BBa_J23119 and BBa_B0015 however appeared sufficient for the protocol to be continued
Miniprep of Samples
(LOGO) Miniprep Protocol ?
The above protocol was done only for J23119 and B0015 – three stocks of each, originating from each of the three replicate falcon tubes
Restriction Digest and Gel Electrophoresis of Samples
(LOGO) Enzyme Digest
Step 2: Set up Reaction 1(Plasmid) and Reaction 2 (Control) for both the miniprepped BioBricks.
| Samples | Recipe | Enzymes | |
|---|---|---|---|
| BioBrick | BBa_J23119 | Digested Plasmid | Xba1 & Spe1 |
| Undigested Plasmid (Control) | None | ||
| BBa_B0015 | Digested Plasmid | Xba1 & Spe1 | |
| Undigested Plasmid (Control) | None | ||
Results: The gel demonstrated no bands for the plasmid and the uncut DNA, which would suggest that there is a low concentration of the plasmid. Therefore we undertook a Nanodrop of the sample.
Nanodrop: (LOGO) Nanodrop
Results
| BioBrick | λ260 | λ 280 |
|---|---|---|
| BBa_J23119 (ng/μl) | 24.8 | 26.1 |
| BBa_B0015 (ng/μl) | 41.7 | 56.6 |
Conclusion: The concentration of these plasmids is very low

Monday 23.7.12
Aim - Transformation of TetR BBa_C0040 BioBrick
Step 1 - Thawing cells: Thaw all materials on ice
Step 2 - Adding Ingredient: Add the following ingredients to autoclaved/sterile eppendorf tubes
| Component | Amount (ul) (one enzyme used) | Amount (ul) (two enzymes used) |
|---|---|---|
| dH20 | 2.5 | 1.5 |
| Buffer 1x | 1 | 1 |
| DNA template | 5 | 5 |
| BSA | 0.5 | 0.5 |
| Enzyme 1 | 1 | 2 |
| Enzyme 2 | N/A | 1 |
Step 3 - Addition of BioBrick: Flick contents gently and centrifuge.
Step 4 - Centrifuge:
RPM: 14000
Time: 1 minute
Temperature: 18oC
Step 5 - Digest Program: Place the samples on a thermocycler under the following conditions:
RPM: 550
Time: 2.5 hours
Temperature: 37oC
Step 6 - Denaturing Enzymes: If you are not running the samples on a gel immediately, denature the restriction enzymes by running the samples on a thermocycler under the following conditions:
RPM: 550
Time: 25 minutes
Temperature: 65oC
Step 1 – Thawing Cells: Use W3100 cell line created in Week 2 (Expt 2.1)
Step 3 – Addition of BioBrick: To a 2ml eppendorf, add 1ul of the following BioBricks. Note: we have changed the protocol for our positive control. Previously it contained no BioBrick, but it has been recommended to us that we transform our positive control such that there is one for each BioBrick – this will tell us if the BioBrick has in any way affected cell viability. This will be used from this point onwards. Include an extra tube as a negative control, with no BioBrick added
| Samples | Function | Module | |
|---|---|---|---|
| BioBrick | BBa_C0040 | Tetracycline Repressor | Buoyancy |
| Control | Positive (Contains BioBrick BBa_C0040) | ||
| Negative (No Biobrick) | |||
Step 9 - Plating samples on Agar Plates: The table below indicates the chosen inoculation volume (two for each BioBrick) and the correct gel antibiotic concentration for all samples.
| Samples | Volume Inoculated | Antibiotic in Gel (ug/ml) | |
|---|---|---|---|
| BioBrick | BBa_C0040 | 10ul | Ampicillin(50ug/ml) |
| 90ul | |||
| Control | Positive (Contains BioBrick BBa_C0040) | 36ul | No Antibiotic |
| Negative (No BioBrick) | 36ul | 2x Ampicillin(50ug/ml) | |
Tuesday 24.7.12
Aim - Results of Transformation
Result: The table below indicates that there was growth for this transformation.
| Samples | Volume Inoculated | Colony Formation | |
|---|---|---|---|
| BioBrick | BBa_C0040 | 10ul | Yes |
| 90ul | Yes | ||
| Control | Positive (Contains BioBrick BBa_C0040) | 36ul | Yes |
| Negative (No BioBrick) | 36ul | No | |

Monday 23.7.12
Aim - Repeat Restriction Digest for BioBricks in Expt 4.1 and 5.1, where the gel previously was inconclusive: This is intended to diagnose whether the correct plasmid had been transformed into our bacteria, by measuring the size of the digested products against a DNA ladder. In previous gel attempts, K540000 has produced a band of the correct size, but we are repeating it because of the presence of other unexpected bands, which we expect are contaminants from the reaction. A previous restriction digest of BBa_I13522 has failed to produce a band of the correct size, so we are repeating the digest before considering another transformation. For the same reason, we are repeating the digest of BBaK398108, which produced bands of incorrect size, which is suggestive of contamination.
(LOGO) Restriction Digest
Step 2: Set up as follows
| Samples | Recipe | Enzymes | |
|---|---|---|---|
| BioBricks | BBa_ K540000 (Expt 4.1) | Digested | Xbar 1 & Spe1 |
| BBa_ I13522 (Expt 4.1) | Digested | Xbar 1 & Spe1 | |
| BBa_ K398108 (Expt 5.1) | Digested | Xbar 1 & Spe1 | |
| BBa_ KI32003 (Expt 5.1) | Digested | Xbar 1 & Spe1 | |
(LOGO) Gel Electrophoresis
Results: The image below shows a 1% Agarose Gel of an Analytical Restriction Enzyme Digest for Expt 7.2. Visible in Lane 1 is a product of the correct size for the BBa_K540000 insert (3123bp), as indicated by A. Also shown is a correct sized product for the backbone PSB1C3 (2070bp), as indicated by B. Lane 2 shows a product corresponding to the size of the BBa_I13522 insert (937bp) as indicated by C. Also present is a product corresponding to the size of the PSB1A3 backbone (2155bp) as indicated by D. Lane 3 has a product corresponding to the expected size of insert BBa_K398108 (844bp) as indicated by E, and a product of the expected size for the plasmid backbone PSB1C3 (2070bp) as indicated by F. Lane 4 shows no products, where we would expect a product of size 1849bp (indicated by G) and a product of 2155 (indicated by H).

Conclusion: Plasmids Bba_k540000 (3123), Bba_I13522 (937) and Bba_K391108 (844) have produced bands of the correct size. The band at 2000 is the plasmid backbone. The failure of Bba_K123003 to produce a band is not of great concern, as we do not expect we will require this BioBrick. However, we may repeat it at a later date.

Tuesday 24.7.12
Aim - Colony Picking from unsuccessful colonies for Expt 7.1 and 6.3
(LOGO) Picking Colonies
Step 2 – Inoculating Colonies into a Selective Broth: The table below indicates the volume of broth and the concentration of antibiotic required for each BioBrick.
| Samples | Number of Falcons | Broth | Antibiotic | |
|---|---|---|---|---|
| Biobrick | BBa_C0040 | 3 | Lysogeny Broth (5ml) | Ampicillin(50ug/ml) |
| BBa_I750016 | 4 | |||
| BBa_B0034 | 5 | |||
| BBa_B0015 | 3 | |||
| BBa_J23119 | 2 | |||
Wednesday 23.7.12
Aim - Results from Colony Picking
Results:
| Samples | Number of Falcons | Growth | |
|---|---|---|---|
| BioBrick | BBa_C0040 | 3 | None |
| BBa_I750016 | 4 | None | |
| BBa_B0034 | 5 | Growth in one | |
| BBa_B0015 | 3 | Growth | |
| BBa_J23119 | 2 | Growth | |
Conclusion: We are beginning to consider other possibilities for the failure of Colony Picking in Expt 7.1 and Expt 6.3. Foremost, we believe it may be due to the addition of Ampicillin to the Agar before it has cooled sufficiently, leading to degradation of the antibiotic. This would reduce the selective pressure, and allow growth of colonies that are not Ampicillin-resistant. Subsequent inoculation into Amipicillin positive LB broth leads to failed growth. Those BioBricks that have worked will undergo Miniprep, Restriction Enzyme Digests and Nanodrop.
Method
Miniprep: (LOGO) Miniprep Protocol 1 (Qiagen)
Step 1: Set up an eppendorf as follows
Step ?: Used 2mls
Step ?: Step ? had to be missed because we realised too late we had too little PE? buffer remaining.
Restriction Digest:
(LOGO) Restriction Digest Protocol 1
Step 2 - Setting up Digests and Controls: The protocol describes the recipe for (i) Digested Plasmid and (ii) Uncut Control. The table below indicates that an uncut and an Xcar1/Spe1 digested sample be set up for each BioBrick.
et up Eppendorfs as follows
| Samples | Recipe | Enzymes | |
|---|---|---|---|
| BioBrick | BBa_B0034 | Digested Plasmid | Xbar 1 & Spe1 |
| Undigested Plasmid (Control) | None | ||
| BBa_B0015 | Digested Plasmid | Xbar 1 & Spe1 | |
| Undigested Plasmid (Control) | None | ||
| BBa_B0034 | Digested Plasmid | Xbar 1 & Spe1 | |
| Undigested Plasmid (Control) | None | ||
(LOGO) Gel Electrophoresis
Results: The image below shows a 1% Agarose Gel of an Analytical Restriction Enzyme Digest for Expt 7.2. Visible in Lane 1 is a band corresponding to the Plasmid Backbone PSB1A2 (2079bp) as show by A, but it is not possible to detect our BBa_J23119 insert on this gel, as it is just 35bp long. Lane 2 shows the uncut plasmid for BBa_J23119 (shown by B) but the size difference between A and B appears to large for Bba_J23119 to have been the insert. This will require further investigation. Lane 3 shows a product 129bp long (indicated by C) which is the correct size for our insert BBa_B0015. A stronger band is also visible, indicated by D, which corresponds to the plasmid backbone PSB1AK3 (3189bp). Lane 4 displays a product which is somewhat larger than the expected size for the uncut plasmid (3318bp) as indicated by E. Lane 5 displays the correct product for the PSB1A2 backbone (2079bp) as indicated by F, but the extremely small size of the Bba_B0034 insert (12bp) means that it cannot be detect on this gel. Lane 6 indicates a band larger than expected, and does not correspond to the size of the uncut plasmid (2091bp) as shown by G.

Conclusion: The BBa_B0015 transformation was a success. However, with regard BBa_J23119, we feel that the difference in the product size of A (PSB1A2 plasmid backbone) and B (uncut plasmid) raises concerns as to whether the insert could be as short as 35bp. However, we must also consider the secondary effects had by the conformation of an uncut plasmid on its migration through the gel. It is possible this is sufficient to misplace the plasmid, such that its position does not represent its size. Even without this possibility, however, it would not be possible to determine whether there was a 35bp difference on such a crude scale of ladder. Instead we will reattempt the Analysis against a 25bp ladder, with the intention of detecting the insert. (See Expt 8.1). We have similar concerns for BBa_B0034, and so this too will be run against a 25bp ladder.
Nanodrop:
(LOGO) Nanodrop
| BioBrick | λ260 | λ 280 |
|---|---|---|
| BBa_J23119 (ng/μl) | 77.6 | 71.8 |
| BBa_B0034 (ng/μl) | 70 | 71.2 |
| BBa_B0015 (ng/μl) | 128.9 | 128 |

Thursday 26.7.12
Aim - Transformation of previously failed BioBricks: BBa_I750016 was previously grown on an Agar Plate (Expt 6.3) but failed to produce growth after colony picking. BBa_C0040 was previously grown on an Agar Plate (Expt 7.1) but also failed to produce growth after colony picking. R0040 failed to produce colonies in Expt 5.1.
Method:
(LOGO) Transformation Protocol 1
Step 1 – Thawing Cells: Use W3100 cell line created in Week 2 (Expt 2.1)
Step 3 – Addition of BioBrick: To each 2ml eppendorf, add 1ul of the following BioBricks. Include an extra tube as a control, with no BioBrick added
| Function | Module | ||
|---|---|---|---|
| BioBrick | BBa_I750016 | Gas Vesicle Polycistronic Cluster | Buoyancy |
| BBa_C0040 | Tetracycline Repressor | Buoyancy | |
| BBa_R0040 | TetR Repressible Promoter | Buoyancy | |
| Control | Positive (one for each of the above BioBricks) | ||
| Negative (No BioBrick) | |||
Step 9 – Plating samples on Agar Plates: The table below indicates the chosen inoculation volume (two for each BioBrick) and the correct gel antibiotic concentration for all samples.(Extra caution was taken to allow agar to cool before adding Ampicillin, in case this is the cause of difficulty).
| Samples | Volume Inoculated | Antibiotic in Gel (ug/ml) | |
|---|---|---|---|
| BioBrick | BBa_I750016 | 10ul | Ampicillin(50ug/ml) |
| 90ul | |||
| BBa_C0040 | 10ul | ||
| 90ul | |||
| BBa_R0040 | 10ul | ||
| 90ul | |||
| Control | Positive (Contains BioBrick BBa_C0040) | 36ul | No Antibiotic |
| Negative (No BioBrick) | 36ul | 1x Ampicillin(50ug/ml) | |
Friday 27.7.12
Aim - Check results of Transformation: The table below indicates whether there was growth on the Agar Plates after Transformation. Included below are images of the Agar Plates for each BioBrick.
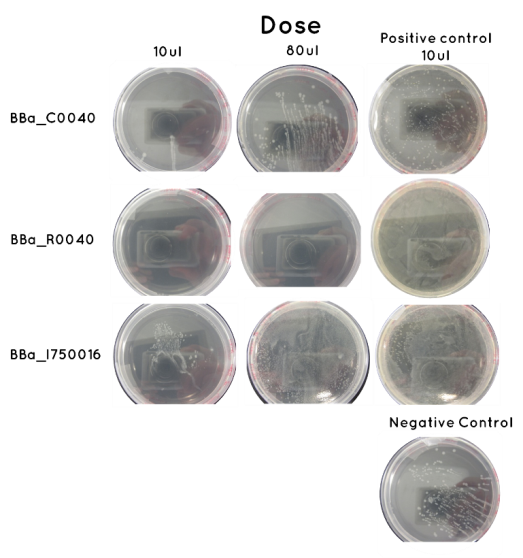
| Samples | Volume Inoculated | Colony Formation | |
|---|---|---|---|
| BioBrick | BBa_I750016 | 10ul | No |
| 90ul | No | ||
| BioBrick | BBa_C0040 | 10ul | Yes |
| 90ul | Yes | ||
| BioBrick | BBa_R0040 | 10ul | Yes |
| 90ul | Yes | ||
| Control | Positive (Contains BioBrick BBa_C0040) | 36ul | Yes |
| Negative (No BioBrick) | 36ul | No | |
Conclusion: Given there was growth on the negative control we are wary of our results. We feel the contamination was not a result of any of our materials, but rather because we were urged to do our colony transfers outside of the sterile hood. This was suggested, because as the inoculation occurs onto an antibiotic positive agar, it should be possible to do the transfer in a non sterile environment. However, this is the first occasion we have had difficulty with out negative control, and so we feel the lack of sterile environment was the cause. Moreover, the pattern of spread on the agar is suggestive of a contaminant. A similar pattern is seen on our BBa_C0040 plates, and so it is likely we will repeat these. A very different pattern was observed on the BBa_I750016 agars, and so we will carry out an analytical digest to determine whether it has been transformed correctly.
 "
"