Team:University College London/Module 3/Modelling
From 2012.igem.org
Module 3: Degradation
Description | Design | Construction | Characterisation | Modelling | Results | Conclusions
Modelling
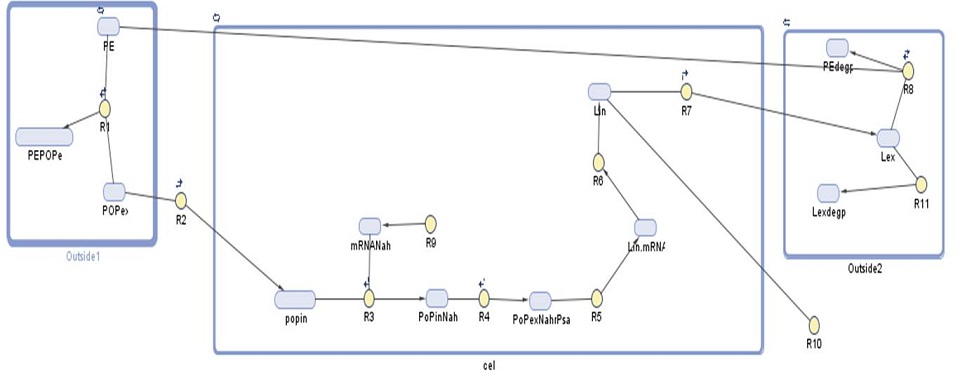
Our gene network model for this module shows the amount of laccase produced by our bacteria. We have used this prediction to find out how many bacteria would be required per cubic meter of sea water in order to effectively degrade polyethylene-based plastics.
| Species | Initial value (molecules) | Notes |
|---|---|---|
| PE | 0.00345 | |
| Lex | 0.0 | |
| PEPOPex | 9.24E-5 | |
| POPin | 0.5 | Dependent on the concentration gradient: when the conc. in is equal to conc. out rate decreases testtestselkjalsfdkjalfkjs;lkdjf;laksjdf;laksjdf;lakjs;dljaf;slkdjfazlskdj;falskjd |
| Nahr | 0.0 | Produced all the time, transcription depends on |
| POPinNahR | 0.0 | Hill function |
| Lin | 0.0 |
| Number | Reaction | Reaction rate (molecules/sec) | Notes |
|---|---|---|---|
| R1 | PE + POPex ↔ PEPOPex | Forward: 1000 Backward: 1 | Pops have 1000 times greater tendency to adhere to plastic than float free in the ocean |
| R2 | POPex ↔ POPin | Forward: 0.6 /n Backward: 0.4 | Based on membrane permeability: diffusion gradient |
| R3 | POPin + mRNA.Nahr → POPin.mRNA.Nahr | Forward: 1 Backward: 0.0001 | The chemical structure/size of POPs is similar to salycilate which is the original compound that react to mRNA.Nahr -> 0 (??) |
| R9 | 0 → mRNA.Nahr | Forward: 0.088 Backward: 0.6 | Transcription rate of NahR in molecules/sec (for NahR size 909 bp2, transcription rate in E.coli 80bp/sec) |
| R4 | POPinmRNANahr → POPinmRNANahr.Psal | Forward: 78200 Backward: 0.191 | |
| R5 | POPexmRNANahr.Psal → Lin.mRNA | 0.054 | Transcription rate of Laccase in molecules/sec (for laccase size 1500 bp1, transcription rate in E.coli 80bp/sec) |
| R6 | Lin.mRNA → Lin | 0.04 | Translation rate of Laccase in molecules/sec (for laccase size 500 aa, translation rate in E.coli 20aa/sec) |
| R11 | Lin → Lindegp | 0.03 | |
| R7 | Lin → Lex | Forward: 0.9 Backward: 0.1 | |
| R8 | Lex → PEdegp | Vm: 0.01 Km: 0.114 |
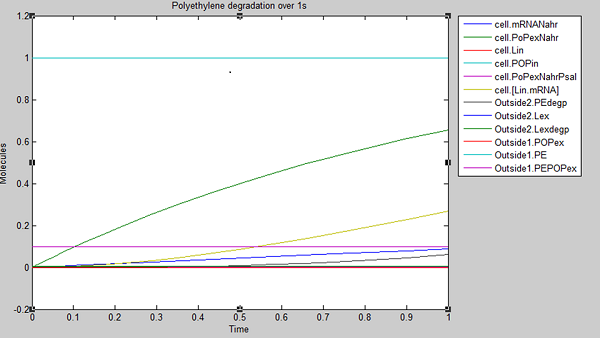
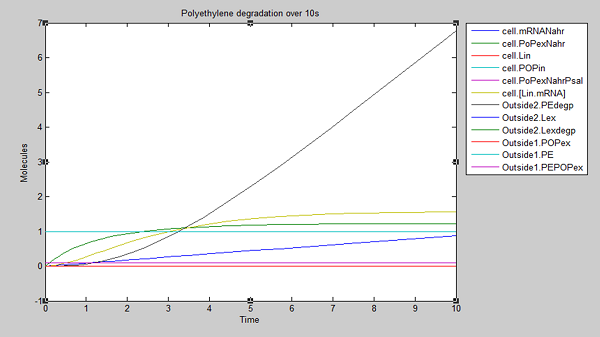
We ran three simulations in SimBiology, each over a different timespan:
1. Laccase size: http://partsregistry.org/Part:BBa_K729002
2. NahR size: http://www.xbase.ac.uk/genome/azoarcus-sp-bh72/NC_008702/azo2419;nahR1/viewer
3. Mato Y, Isobe T, Takada H, Kanehiro H, Ohtake C, Kaminuma T (2001) Plastic Resin Pellets as a Transport Medium for Toxic Chemicals in the Marine Environment. Environ. Sci. Technol. 35: 318-324
4. Teuten E, Saquing J, Knappe D et al. (2009) Transport and release of chemicals from plastics to the environment and to wildlife. Philosophical transactions of the Royal Society of London 364: 2027-2045
8. Young K, Silver LL (1991) Leakage of periplasmic enzymes from envA1 strains of Escherichia coli. J Bacteriol. 173: 3609–3614
11.Van A, Rochman C, Flores E, Hill K, Vargas E, Vargas S, Hoh E (2012) Persistent organic pollutants in plastic marine debris found on beaches in San Diego, California. Chemosphere 86: 258-263
12. Santo M, Weitsman R, Sivan A (2012) The role of the copper-binding enzyme - laccase - in the biodegradation of polyethylene by the actinomycete Rhodococcus ruber. International Biodeterioration & Biodegradation 208: 1-7
 "
"