Team:University College London/LabBook/Week8
From 2012.igem.org
Rwilkinson (Talk | contribs) (→Monday 30.6.12) |
Rwilkinson (Talk | contribs) (→Monday 30.6.12) |
||
| Line 9: | Line 9: | ||
<html><div class="protocol protocol-RestrictionEnzymeDigest">Restriction Enzyme Digest Protocol 1</div><div class="protocolContent"></html>{{:Team:University_College_London/Protocols/RestrictionEnzymeDigest1}}<html></div></html> | <html><div class="protocol protocol-RestrictionEnzymeDigest">Restriction Enzyme Digest Protocol 1</div><div class="protocolContent"></html>{{:Team:University_College_London/Protocols/RestrictionEnzymeDigest1}}<html></div></html> | ||
| + | |||
| + | '''Step 2 - Setting up Digests and Controls:''' The protocol describes the recipe for (i) Digested Plasmid and (ii) Uncut Control. The table below indicates that an uncut and an Xba1/Spe1 digested sample be set up for each BioBrick. Set up Eppendorfs as follows | ||
| + | |||
| + | |||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | ! colspan="2" | Samples !!Recipe !! Enzymes | ||
| + | |- | ||
| + | | rowspan="7" |BioBrick || rowspan="2" | BBa_Bj23119 ||Digested Plasmid || Xba1 & Spe1 | ||
| + | |- | ||
| + | | Undigested Plasmid (Control) || None | ||
| + | |- | ||
| + | | rowspan="2" | BBa_B0034 || Digested Plasmid|| Xba1 & Spe1 | ||
| + | |- | ||
| + | | Undigested Plasmid (Control) || None | ||
| + | |- | ||
| + | |} | ||
| + | |||
<html><div class="protocol protocol-Electrophoresis">Electrophoresis Protocol</div><div class="protocolContent"></html>{{:Team:University_College_London/Protocols/Electrophoresis}}<html></div></html> | <html><div class="protocol protocol-Electrophoresis">Electrophoresis Protocol</div><div class="protocolContent"></html>{{:Team:University_College_London/Protocols/Electrophoresis}}<html></div></html> | ||
Revision as of 08:49, 7 August 2012



Contents |
Monday 30.6.12
Aim: Repeat the gel from Expt 7.3 (week 7) with a smaller ladder and higher agarose percentage in order to detect the very small inserts of BBa_J23119 (35bp) and BBa_B0034 (12hp). Previous ladder used was 10037 bp, and this time we used a 25bp ladder.
Step 1 - Thawing cells: Thaw all materials on ice
Step 2 - Adding Ingredient: Add the following ingredients to autoclaved/sterile eppendorf tubes
| Component | Amount (ul) (one enzyme used) | Amount (ul) (two enzymes used) |
|---|---|---|
| dH20 | 2.5 | 1.5 |
| Buffer 1x | 1 | 1 |
| DNA template | 5 | 5 |
| BSA | 0.5 | 0.5 |
| Enzyme 1 | 1 | 2 |
| Enzyme 2 | N/A | 1 |
Step 3 - Addition of BioBrick: Flick contents gently and centrifuge.
Step 4 - Centrifuge:
RPM: 14000
Time: 1 minute
Temperature: 18oC
Step 5 - Digest Program: Place the samples on a thermocycler under the following conditions:
RPM: 550
Time: 2.5 hours
Temperature: 37oC
Step 6 - Denaturing Enzymes: If you are not running the samples on a gel immediately, denature the restriction enzymes by running the samples on a thermocycler under the following conditions:
RPM: 550
Time: 25 minutes
Temperature: 65oC
Step 2 - Setting up Digests and Controls: The protocol describes the recipe for (i) Digested Plasmid and (ii) Uncut Control. The table below indicates that an uncut and an Xba1/Spe1 digested sample be set up for each BioBrick. Set up Eppendorfs as follows
| Samples | Recipe | Enzymes | |
|---|---|---|---|
| BioBrick | BBa_Bj23119 | Digested Plasmid | Xba1 & Spe1 |
| Undigested Plasmid (Control) | None | ||
| BBa_B0034 | Digested Plasmid | Xba1 & Spe1 | |
| Undigested Plasmid (Control) | None | ||
Preparing the Gel
Step 1: Within a conical flask, add 3ml 50X TAE, 1.5g Agarose, and 150ml RO water
Step 2: Heat for 1 min in microwave. Swirl. Heat again for 30s. If solution is clear stop. Else repeat.
Step 3: Cool solution under running cold water.
Step 4: Add 20ul ethidium bromide (normal concentration of EB solution is 500ug/ul)
Step 5: Pour into a sealed casting tray in a slow steady stream, ensuring there are no bubbles
Running a gel
Step 6: Add 1 part loading buffer to five parts of loading sample
Step 7: Position the gel in the tank and add TAE buffer, enough to cover the gel by several mm
Step 8: Add 5ul of DNA ladder to lane 1
Step 9: Add samples to the remaining wells
Step 10: Run at 100 volts for 1hour and 15 minutes
Imaging the Gel
Step 11: Place gel in GelDoc 2000 chamber
Step 12: Turn GelDoc 2000 chamber on
Step 13: From computer: Quantity One > Scanner > Gel_Doc_Xr>Manuqal Acquire
Step 14: Alter the exposure/settings to give a clear image.
TAE - Tris-acetate-EDTA
EDTA - ethylenediamine tetraacetic acid
Results:The image below shows a 3.5% Agarose Gel of an Analytical Restriction Enzyme Digest for Expt 7.3. The high agarose concentration of the gel was intended to slow the progression of DNA fragments to enable us to detect the small inserts of BBa_J23119 and BBa_B0034. The table below indicates the expected sizes of BioBricks and Plasmid Backbones. Accordingly, we used a the smallest available ladder, with fragments ranging from 25bp to 500bp. At the top of the gel it is possible to see the large fragments we detected on a 1000bp gel in Expt 7.3 Week 7. However, we had expected this gel to also demonstrate the BBa_J23119 insert (35bp) corresponding to letter A in Lane 1, and the BBa_B0034 insert, corresponding to letter B in Lane 3. No product was found. As expected, there was no product measurable against the 25bp ladder for the uncut plasmids in Lane 2 and Lane 4.
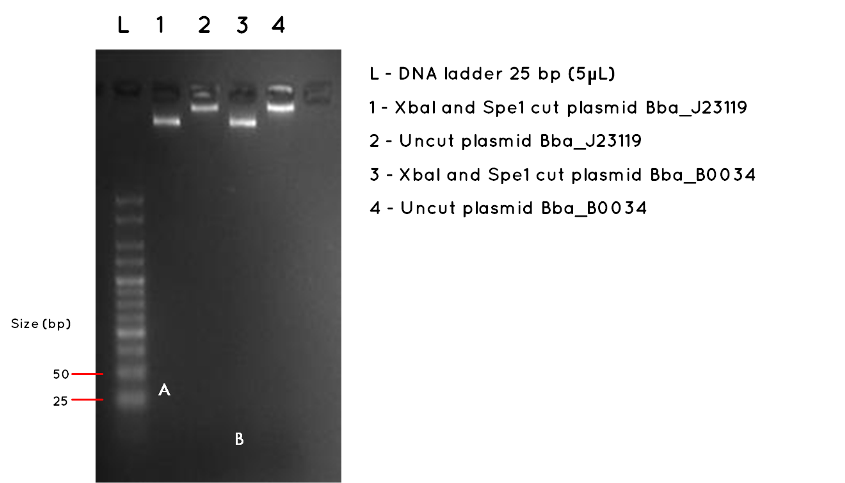
| BioBrick | Expected BioBrick Size (bp) | Plasmid Backbone | Plasmid Expected Size (bp) | Combined Size |
|---|---|---|---|---|
| BBa_J23119 | 35 | PSB1A2 | 2079 | 2114 |
| BBa_B0034 | 12 | PSB1A2 | 2079 | 2091 |
Conclusion:No product was detected for either BioBrick insert. However, we do no feel this necessarily proves the transformation failed, especially as we detected the correct plasmid backbone for each in week 7 (Expt 7.3). The modest nanodrop concentration obtained last week suggests that the concentration was not high enough to allow such a small fragment to be visualised. We are considering alternatives such as running a polyacrylamide gel, which is more sensitive to small fragments, or amplifying the fragment using PCR

7-4

8-1

8-2

8-3

8-4

8-5
 "
"