Team:University College London/LabBook/Week13
From 2012.igem.org
(→Monday 03/09) |
Sednanalien (Talk | contribs) |
||
| Line 68: | Line 68: | ||
| - | + | ||
| - | + | Date Time Colony diameter Halo diameter Absorbance at OD600 | |
| - | + | 03.09.2012 12.30pm 0 0 0 | |
| - | + | 03.09.2012 16.30pm 0 0 0 | |
| - | + | 03.09.2012 19.30 pm 0 0 0 | |
| - | + | 03.09.2012 22.30 pm 0.5mm 1mm 0.002 | |
| - | + | 04.09.2012 01.30 am 1mm 3.5mm 0.182 | |
| - | + | 04.09.2012 04.30 am 1.5mm 7mm 0.238 | |
| - | + | 04.09.2012 07.30 am 2mm 9mm 0.297 | |
| - | + | 04.09.2012 10.30am 2.5mm 11mm 0.518 | |
| - | + | 04.09.2012 12.30pm 3mm 12.5mm 0.701 | |
| - | + | 04.09.2012 14.30pm 3.5mm 14mm 0.811 | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
04.09.2012 16.30pm 4mm 15mm 0.906 | 04.09.2012 16.30pm 4mm 15mm 0.906 | ||
Revision as of 03:17, 27 September 2012



Contents |
Monday 03/09
Aim:
To repeat the DNAse/Nuclease Halo experiment in order to check whether results are repeatable. This included collecting data of halo diameters and colony diameters over 28 hour, and to determine how the nuclease production increases overtime.
Methods:
In order to be able to characterise cells, the following protocol was used:
1. Prepare 11-16 plates (10ml DNAse - Agar + 10ul AMP + 10ul 1M IPTG). IPTG induces the lac promoter which in turn activates the transcription of nuclease.
2. Streak cells onto all plates at the same time
3. Incubate at 37°C
4. Apply hydrochloric acid (HCl) to the first plate before putting in the incubator (set time as zero)
5. Take a second reading after four hours, followed by six readings every 3 hours, and a final three readings every two hours.
6. When the reading is taken, observe the following:
a) Diameter of the colony (once the diameter of the colony is measured, pick the colony and put it to grow in LB for nine hours)
b) Diameter of the halo that is achieved once HCl is applied
c) OD from a)
d) Estimate of the depth of the colony on the agar plate
For BL21 cell line that has been modified to contain nuclease:
1. Prepare 11-16 plates (DNAse - Agar + CMP)
2. Streak cells onto all plates at the same time
3. Incubate at 37°C
4. Apply HCl to the first plate before putting in the incubator (set time as zero)
5. Take a second reading after four hours, followed by six readings every 3 hours, and a final three readings
every two hours.
6. When the reading is taken, observe the following:
a. Diameter of the colony (once the diameter of the colony is measured, pick the colony and put it to grow in LB for nine hours)
b. Diameter of the halo that is achieved once HCL is applied
c. OD from a)
d. Estimate of the depth of the colony on the agar plate
Results:
The following table images show the results of the DNase assay:
| No | Part | Enzymes |
|---|---|---|
| 1 | laccase | X+P |
| 2 | curli | X+P |
| 3 | irrE | X+P |
| 4 | irrE | E+P |
| 5 | nuclease | X+P |
| 6 | nuclease | X+P |
| 7 | plasmid backbone | E+P |
| 8 | Plasmid backbone | E+P |
| 9 | Linker | E+S |
Step 3 - Aim:
To carry out ligations using the inserts and backbone prepared previously. The ligations carried out included qoui???? Vat iz zis writing down here?
X+P cut to E+P cut Plasmid All.
X+S Plasmid backbone religated control and no ligase control.
E+P cut ire to E+P cut plasmid backbone.
Methods:
Quick T4 Ligase was used to carry out ligations. The protocol followed in order to carry out ligations was as supplied with the reagants. Do we need to write it down?
Step 4 - Aim:
To carry out a transformation of the ligation
Methods:
Protocol? 5uL used to transform 100uL cells.
Thursday 06/09
Aim:
To check whether transformations using ligation product on the previous day were successful.
Results:
No colonies were visible on the plates. Hence, the transformation was considered to be unsuccessful and needs to be repeated.

Wednesday 05/09
Aim:
To characterise the two strains of laccase obtained from the laccase ligation.
Method:
1) W3110 cells transformed with laccase and control W3110 cells are inoculated in 10mL of LB media, and incubated overnight at 37˚C in a 200rpm shaker
2) The O/N culture is then centrifuged at 6100g for 20 minutes, in order to extract the media containing laccase. The supernatant is retained.
3) In order to take readings, cuvettes are prepared with 2.2mL KH2PO4 buffer + 0.5mL Laccase supernatant + 0.3mL Syringaldazine, which is added immediately before readings are begun (as explained in step 4). A blank is created using 0.5mL of LB media instead of laccase supernatant.
4) The optical density at 530nm of each of the samples is measured at five minute intervals over 30 minutes.
Result:
Not shown
Conclusion:
We have determined that laccase strain 2 is a better laccase producing strain. Furthermore, we have proven the function of our BioBrick, as the oxidation activity of both laccase stratins was significantly higher than that of the control W3110 strain.

Thursday 06/09
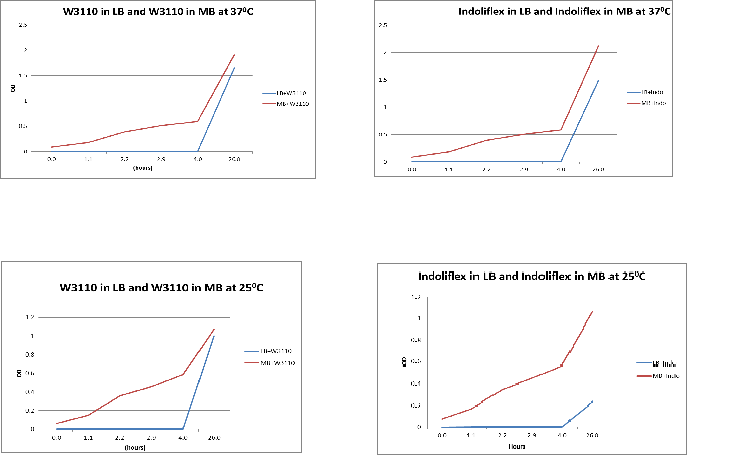
Aim:
To compare the difference in growth of Indoli and E. coli growth in LB & MB media
Methods:
1. Inoculate 10ul of W3110 glycerol stock in 10ml of LB and another 10ul of W3110 glycerol stock in 10ml of MB
2. Repeat this with an indolifex glycerol stock
3. Measure the absorbence of all four samples every hour for 12 hours. Then take a final measurement after another 12 hours. Thus, measurements span 24 hours.
Results:
The following graphs show growth of the different strains and different media. The flasks were placed in a 370C shaker at 200rpm.
Conclusion:
From the above graphs, it can be seen that marine bacteria grows better in MB, irrespective of the temperature used.
 "
"