Team:University College London/LabBook/Week14
From 2012.igem.org
YanikaBorg (Talk | contribs) (→Monday 10.09.12) |
YanikaBorg (Talk | contribs) (→Friday 14.09.12) |
||
| Line 105: | Line 105: | ||
|- | |- | ||
|BBa_K729001 & BBA_J23100 || 933 & 35 || 968 || pSB1C3 || 2070 || 3038 | |BBa_K729001 & BBA_J23100 || 933 & 35 || 968 || pSB1C3 || 2070 || 3038 | ||
| + | |- | ||
| + | |BBa_K729001 & BBA_J23106 || 933 & 35 || 968 || pSB1C3 || 2070 || 3038 | ||
|- | |- | ||
|BBa_K729001 & BBa_J23119 & BBa_B0034 || 933 & 35 & 12 || 980 || pSB1C3 || 2070 || 3050 | |BBa_K729001 & BBa_J23119 & BBa_B0034 || 933 & 35 & 12 || 980 || pSB1C3 || 2070 || 3050 | ||
Revision as of 01:35, 27 September 2012



Contents |
Sunday 09.09.12
Aim: Day 1 of Generating competent cells.
Method:
Step 1 - Addition of BioBrick: To the still frozen competent cells, add 1 - 5 µL of the resuspended DNA to the 2ml tube.
Step 4 - Incubation: Close tube and incubate the cells on ice for 45 minutes.
Step 5 - Heat Shock: Heat shock the cells by immersion in a pre-heated water bath at 37ºC for 10 minutes.
Step 6 - Incubation: Incubate the cells on ice for 2 minutes.
Step 7 - Add media: Add 1.5ml of Lysogeny Broth and transfer to a falcon tube.
Step 8 - Incubation: Incubate the cells at 37ºC for 1 hour at RPM 550.
Step 9 - Transfer: transfer the solution back into a 1.5ml Eppendorf and centrifuge
RPM: 14000
Time: 2 minutes
Temperature (18-25oC)
Step 10 - Resuspend:Remove supernatant and resuspend in 100ul LB
Step 11 - Plating: Spread the resuspended cell solution onto a selective nutrient agar plate. Place the plates in a 37°C static incubator, leave overnight (alternatively a 30°C static incubator over the weekend)
Monday 10.09.12
Aim: Day 2 of Generating competent cells. The aim is to incubate cells in the presence of CaCl2 to prepare the cell wall, allowing it to become permeable to DNA for transformation.
Method:
Step 1 -Creating culture media: In a sterile environment, set up a falcons, with 5mls of Lysogeny Broth and 100ul 1M MgS04.
Step 2 – Selecting a Colony: Select a clear, isolated colony and using an inoculation hoop, scoop up a colony onto the tip. Deposit in the falcon tube
Step 3 - Cell culture: Culture your falcon tubes overnight at a temperature of 37 oC. Leave for no longer than 16 hours.
Tuesday 11.09.12
Aim: Day 3 of Generating competent cells
Method:
Step 1 – Innoculation: Inoculate 100mls of Lysogeny Broth in pre-warmed conical with 1ml of the overnight culture from Day 2
Step 2 – Incubation: Incubate for two hours in a 37oC shaker until the cells are at the early log phase of the growth curve. Measure absorbance on a spectrometer until A6000 is approximately 0.3.
Step 3 – Incubation: Transfer to a chilled sterile 50ml Falcon tube and incubate on ice for 10 minutes Step 4 – Centrifuge:
-Time: 5 mins -G – 3300 -Temperature: 180C
Step 5 – Incubate: Resus in 10ml of ice cold 0.1M CaCl2 in 15% glycerol and incubate on ice for 30 minutes.
Step 6 – Centrifuge: -Time: 5 mins -G – 3300 -Temperature: 180C
Step 7 – Transfer: Resus in 1ml of ice cold 0.1M CaCl2 in 15% glycerol. Transfer 100ul aliquots to pre-chilled, pre-labelled eppendorf tubes. Store at -70oC
Step 8 – Centrifuge -Time: 5 mins -G – 3300 -Temperature: 180C
Step 9 - Storage: Store cells at -80oC

Wednesday 12.09.12
Aim: To carry out a transformation using the 3A ligation products in order to be able to analyse whether ligation was successful.
Method:
Step 1 - Addition of BioBrick: To the still frozen competent cells, add 1 - 5 µL of the resuspended DNA to the 2ml tube.
Step 4 - Incubation: Close tube and incubate the cells on ice for 45 minutes.
Step 5 - Heat Shock: Heat shock the cells by immersion in a pre-heated water bath at 37ºC for 10 minutes.
Step 6 - Incubation: Incubate the cells on ice for 2 minutes.
Step 7 - Add media: Add 1.5ml of Lysogeny Broth and transfer to a falcon tube.
Step 8 - Incubation: Incubate the cells at 37ºC for 1 hour at RPM 550.
Step 9 - Transfer: transfer the solution back into a 1.5ml Eppendorf and centrifuge
RPM: 14000
Time: 2 minutes
Temperature (18-25oC)
Step 10 - Resuspend:Remove supernatant and resuspend in 100ul LB
Step 11 - Plating: Spread the resuspended cell solution onto a selective nutrient agar plate. Place the plates in a 37°C static incubator, leave overnight (alternatively a 30°C static incubator over the weekend)
Thursday 13.09.12
Aim:
Picking of colonies from the ligation transformation plates
Methods:
Step 1 - Picking Colony: Using a pipette select an isolated colony
Step 2 - Deposit: Dip the colony in 10ul of dH20, preferably in screw top container
Step 3 - Boiling Stage:Boil the dH20 100 for 5-10mins
Step 4 - Centrifuge:
Time: 1 min
RPM: 8000
Temp: 18oC
Step 5 - Remove Supernatant: The supernatant contains the DNA template.
Friday 14.09.12
Aim:
To check whether the 3A assembly ligation has been successful by purifying the overnight culture from the previous day, carrying out an analytical digest and analysing results on a gel.
Methods:
A mini-prep was carried out as per the following protocol:
This was followed by a 10ul analytical digest as per the following:
Step 2 - Adding Ingredient: Add the following ingredients to autoclaved/sterile eppendorf tubes
Insert Table
Step 3 - Addition of BioBrick: Flick contents gently and centrifuge.
Step 4 - Centrifuge:
RPM: 14000
Time: 1 minute
Temperature: 18oC
Step 5 - Digest Program: Place the samples on a thermocycler under the following conditions:
RPM: 550
Time: 1 hour
Temperature: 37oC
Step 6 - Denaturing Enzymes: If you are not running the samples on a gel immediately, denature the restriction enzymes by running the samples on a thermocycler under the following conditions:
RPM: 550
Time: 25 minutes
Temperature: 65oC
Temperature: 18oC
Finally, results of the figest were run on a gel as follows:
Preparing the Gel
Step 1: Within a conical flask, add 3ml 50X TAE, 1.5g Agarose, and 150ml RO water
Step 2: Heat for 1 min in microwave. Swirl. Heat again for 30s. If solution is clear stop. Else repeat.
Step 3: Cool solution under running cold water.
Step 4: Add 20ul ethidium bromide (normal concentration of EB solution is 500ug/ul)
Step 5: Pour into a sealed casting tray in a slow steady stream, ensuring there are no bubbles
Running a gel
Step 6: Add 1 part loading buffer to five parts of loading sample
Step 7: Position the gel in the tank and add TAE buffer, enough to cover the gel by several mm
Step 8: Add 5ul of DNA ladder to lane 1
Step 9: Add samples to the remaining wells
Step 10: Run at 100 volts for 1hour and 15 minutes
Imaging the Gel
Step 11: Place gel in GelDoc 2000 chamber
Step 12: Turn GelDoc 2000 chamber on
Step 13: From computer: Quantity One > Scanner > Gel_Doc_Xr>Manuqal Acquire
Step 14: Alter the exposure/settings to give a clear image.
TAE - Tris-acetate-EDTA
EDTA - ethylenediamine tetraacetic acid
Results:
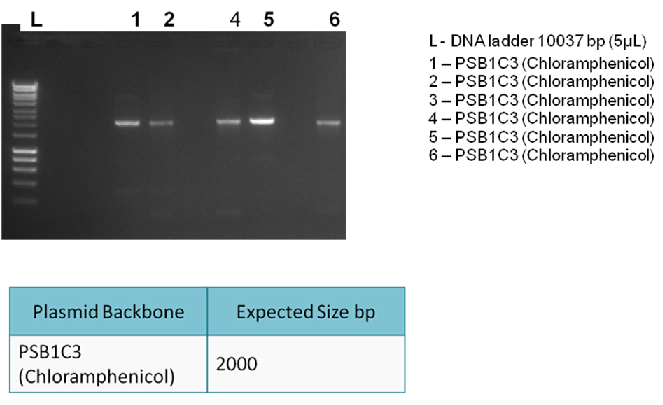
The following digram shows results of the analytical digest, with each purified DNA being cut once and twice in order to confirm identity:
| BioBricks | BioBrick Size (bp) | Ligation size (bp) | Plasmid Backbone | Plasmid Expected Size (bp) | Combined Size |
|---|---|---|---|---|---|
| BBa_K729001 & BBA_J23100 | 933 & 35 | 968 | pSB1C3 | 2070 | 3038 |
| BBa_K729001 & BBA_J23106 | 933 & 35 | 968 | pSB1C3 | 2070 | 3038 |
| BBa_K729001 & BBa_J23119 & BBa_B0034 | 933 & 35 & 12 | 980 | pSB1C3 | 2070 | 3050 |
| BBa_K729002 & BBa_J23119 & BBa_B0034 | 484 & 35 & 12 | 531 | pSB1C3 | 2070 | 2601 |
Conclusion:
The bands on the gel indicate are not as expected. Hence, it is concluded that the ligation was unsuccessful.

Tuesday 11.09.12
Step 1 - Aim:
To generate enough plasmid backbone (pSB1C3) to be used in a 3A ligation. This is followed by a PCR clean-up.
Methods:
Step 1 - Setting up PCR tubes: Thaw reagents and add to PCR tubes in the proportions described in the table below
| PCR Components | Volume (ul) |
|---|---|
| 5x Reaction Buffer | 10 |
| 25mM MgCl2 | 4 |
| 10mM dNTPs | 1 |
| 10uM Forward primer | 5 |
| 10uM Reverse primer | 5 |
| DNA Polymerase | 0.25 |
| Nuclease Free Water | 24.25 |
| Template DNA | 0.5 |
| Total Volume | 0.5 |
Step 2 - PCR program: Add PCR tubes to a thermocycler and run under the following conditions.
| PCR conditions | Temp (oC) | Time (s) |
|---|---|---|
| Initial Denaturation (1 cycle) | 95 | 30 |
| Denaturation/Annealing/Extension (30 cycles) | 95/55/72 | 10/25/120 |
| Final Extension (1 cycle) | 72 | 600 |
| Hold | 4 | ∞ |
Step 2 - Aim:
To check whether PCR is successful. After the PCR reaction, we can detect the presence of the correct products by running a 1% gel electrophoresis.
Methods:
Preparing the Gel
Step 1: Within a conical flask, add 3ml 50X TAE, 1.5g Agarose, and 150ml RO water
Step 2: Heat for 1 min in microwave. Swirl. Heat again for 30s. If solution is clear stop. Else repeat.
Step 3: Cool solution under running cold water.
Step 4: Add 20ul ethidium bromide (normal concentration of EB solution is 500ug/ul)
Step 5: Pour into a sealed casting tray in a slow steady stream, ensuring there are no bubbles
Running a gel
Step 6: Add 1 part loading buffer to five parts of loading sample
Step 7: Position the gel in the tank and add TAE buffer, enough to cover the gel by several mm
Step 8: Add 5ul of DNA ladder to lane 1
Step 9: Add samples to the remaining wells
Step 10: Run at 100 volts for 1hour and 15 minutes
Imaging the Gel
Step 11: Place gel in GelDoc 2000 chamber
Step 12: Turn GelDoc 2000 chamber on
Step 13: From computer: Quantity One > Scanner > Gel_Doc_Xr>Manuqal Acquire
Step 14: Alter the exposure/settings to give a clear image.
TAE - Tris-acetate-EDTA
EDTA - ethylenediamine tetraacetic acid
Results:
The following gel results indicate whether PCR was successful.
Conclusion:
Since band sizes were as expected, the PCR was considered to be successful. Hence, the backbone was purified and used in the 3A ligation.

Thursday 13.09.12
Aim:
To generate enough linker (BBa_J23119 + BBa_B0034) to be used in a 3A ligation. This is followed by a PCR clean-up.
Methods:
Step 1 - Setting up PCR tubes: Thaw reagents and add to PCR tubes in the proportions described in the table below
| PCR Components | Volume (ul) |
|---|---|
| 5x Reaction Buffer | 10 |
| 25mM MgCl2 | 4 |
| 10mM dNTPs | 1 |
| 10uM Forward primer | 5 |
| 10uM Reverse primer | 5 |
| DNA Polymerase | 0.25 |
| Nuclease Free Water | 24.25 |
| Template DNA | 0.5 |
| Total Volume | 0.5 |
Step 2 - PCR program: Add PCR tubes to a thermocycler and run under the following conditions.
| PCR conditions | Temp (oC) | Time (s) |
|---|---|---|
| Initial Denaturation (1 cycle) | 95 | 30 |
| Denaturation/Annealing/Extension (30 cycles) | 95/55/72 | 10/25/120 |
| Final Extension (1 cycle) | 72 | 600 |
| Hold | 4 | ∞ |
Preparing the Gel
Step 1: Within a conical flask, add 3ml 50X TAE, 1.5g Agarose, and 150ml RO water
Step 2: Heat for 1 min in microwave. Swirl. Heat again for 30s. If solution is clear stop. Else repeat.
Step 3: Cool solution under running cold water.
Step 4: Add 20ul ethidium bromide (normal concentration of EB solution is 500ug/ul)
Step 5: Pour into a sealed casting tray in a slow steady stream, ensuring there are no bubbles
Running a gel
Step 6: Add 1 part loading buffer to five parts of loading sample
Step 7: Position the gel in the tank and add TAE buffer, enough to cover the gel by several mm
Step 8: Add 5ul of DNA ladder to lane 1
Step 9: Add samples to the remaining wells
Step 10: Run at 100 volts for 1hour and 15 minutes
Imaging the Gel
Step 11: Place gel in GelDoc 2000 chamber
Step 12: Turn GelDoc 2000 chamber on
Step 13: From computer: Quantity One > Scanner > Gel_Doc_Xr>Manuqal Acquire
Step 14: Alter the exposure/settings to give a clear image.
TAE - Tris-acetate-EDTA
EDTA - ethylenediamine tetraacetic acid

Friday 14.09.13
Aim:
To characterise nuclease activity BL21 cells
Method:
1. Prepare 11-16 plates (10ml LBAgar+10ul CMP )
2. Streak cells onto all plates at the same time
3. Incubate at 37°C
4. Apply hydrochloric acid (HCL) to the first plate before putting them in the incubator (set time as zero)
5. Take a second reading after four hours, followed by six readings every 3 hours, and a final three readings every two hours. When the reading is taken, observe the following:
a) Diameter of the colony (once the diameter of the colony is measured, pick the colony and put it to grow in LB for nine hours)
b) Diameter of the halo that is achieved once HCL is applied
c) OD from a)
d) Estimate of the depth of the colony on the agar plate
Results:
The following table shows readings of the diameters and OD at different time points:
| Date | Time | Colony diameter/mm | Halo diameter/mm | Absorbance at 600 OD |
|---|---|---|---|---|
| Friday | 12.30 | 0 | 0 | 0 |
| Friday | 16.30 | 0 | 0 | 0 |
| Friday | 19.30 | 0 | 0 | 0 |
| Friday | 22.30 | 0 | 0 | 0 |
| Saturday | 01.30 | 0.5 | 1.5 | 0.068 |
| Saturday | 04.30 | 1 | 2 | 0.098 |
| Saturday | 07.30 | 1.5 | 3 | 0.159 |
| Saturday | 10.30 | 1.5 | 3.5 | 0.192 |
| Saturday | 12.30pm | 2 | 4 | 0.203 |
| Saturday | 14.30 | 2 | 4 | 0.209 |
| Saturday | 16.30 | 2.5 | 5 | 0.215 |
The average depth of the colonies was noted to be 0.5 - 1mm
Conclusion:
It can be seen that nuclease works as expected in BL21 cells. However, nuclease activity is less efficient when compared to nuclease activity in WNu cells. This is in line with expectations, since nuclease is found naturally in WNu cells but not in BL21 cells.
 "
"