Team:Goettingen/Notebook/Results
From 2012.igem.org
| Line 1: | Line 1: | ||
{{GoettingenHeader|deu=Team:Goettingen/Notebook/Results_deu|eng=Team:Goettingen/Notebook/Results}} | {{GoettingenHeader|deu=Team:Goettingen/Notebook/Results_deu|eng=Team:Goettingen/Notebook/Results}} | ||
| - | + | __TOC__ | |
== Homing coli: Engineering <i>E. coli</i> to become tracking dogs == | == Homing coli: Engineering <i>E. coli</i> to become tracking dogs == | ||
The model organism <i>Escherichia coli</i> is naturally capable of sensing substances in its environment and consequently moves directionally | The model organism <i>Escherichia coli</i> is naturally capable of sensing substances in its environment and consequently moves directionally | ||
| Line 18: | Line 18: | ||
== #3 - Chemoreceptor Library == | == #3 - Chemoreceptor Library == | ||
| - | [[File: | + | [[File:Gruppe3 Figure01.png|500px|thumb|center| <b>Figure 1: Library design of the quik changed chemoreceptor Tar. </b> <b>(A)</b> Schematic representation of the applied procedure for library generation of the quik-changed Tar_QC under a strong promoter. Saturated PCR mutagenesis was performed in two rounds. In the first step the plasmid pSB1C3_18C_TAR_QC was amplified with primers that randomly mutate three triplets and introduce a <i>Bsa</i>I restriction site (scarless mutagenesis). In the second step the parent plasmid was digested with DpnI and the mutated plasmid with <i>Bsa</i>I to provide sticky ends. <i>Bsa</i>I cuts outside the restriction site. In step three the plasmid was ligated. Step four includes the transformation of the mutated DNA into electrocompetent DH10B cells. Subsequently the DNA was isolated for the second round of mutagenesis PCR (step five), where two other triplets were mutagenized repeating the previous steps. The 1<sup>st</sup> and 2<sup>nd</sup> round of the mutation library covered 10<sup>5</sup> clones. The two rounds were combined to one library with a final diversity of 2x 10<sup>5</sup>. <b>(B)</b> Representative pictures of 0.8% Agarose gels from the first saturated PCR mutagenesis round of step two after PCR (2), step three after double digest with <i>Dpn</i>I and <i>Bsa</i>I (3) and step four after ligation of the mutant plasmid (4). The size of the plasmid is always 3769 bp. As standard a 1kb Molecular Weight Marker (M; Fermentas) was used. <b>(C)</b> Overview (right) and detailed (left) PYMOL images of Tar with aspartate in its binding pocket (PDB ID: 1WAT). The mutated amino acid residues involved in ligand binding are labeled. <b>(D)</b> Alignment of the regions involved in ligand binding of the original DNA sequence of Tar_QC (pSB1C3_TAR_QC), the library sequence (TAR_QC Library) and the amino acid sequence (TAR_QC aa-sequence). The upper two lines show the site of mutation, here. K stands for guanine or thymine, N for any possible base. Corresponding to the DNA sequence the amino acid sequence is shown with labeled positions of mutated amino acid residues (*) above. G: guanine, C: cytosine, A: adenine, T: thymine.]] |
| + | |||
| + | [[File:Gruppe3 Figure02.png|500px|thumb|center| <b>Figure 2: Characteristics of the generated chemoreceptor Tar_QC library. </b> <b>(A)</b> Representation of the reached diversity in the two rounds of Tar_QC library generation. Both, 1<sup>st</sup> and 2<sup>nd</sup> round, yielded 10<sup>5</sup> clones. After combination of the two diversities the merged library resembles 2x 10<sup>5</sup> clones. <b>(B)</b> Sequencing chromatogram excerpt of the generated chemoreceptor Tar_QC library in both rounds.Sequencing results of the 1<sup>st</sup> round (top)with mutations for the amino acidsat position 149, 150 and 154 and 2<sup>nd</sup> round (bottom) with mutations at 69 and 73. Inserted mutations are indicated by the triplets consisting of N and K, whereby K stands for guanine or thymine, N for any possible base. Only in the mutated regions no distinct peaks are present.G: guanine, C: cytosine, A: adenine, T: thymine.]] | ||
| + | |||
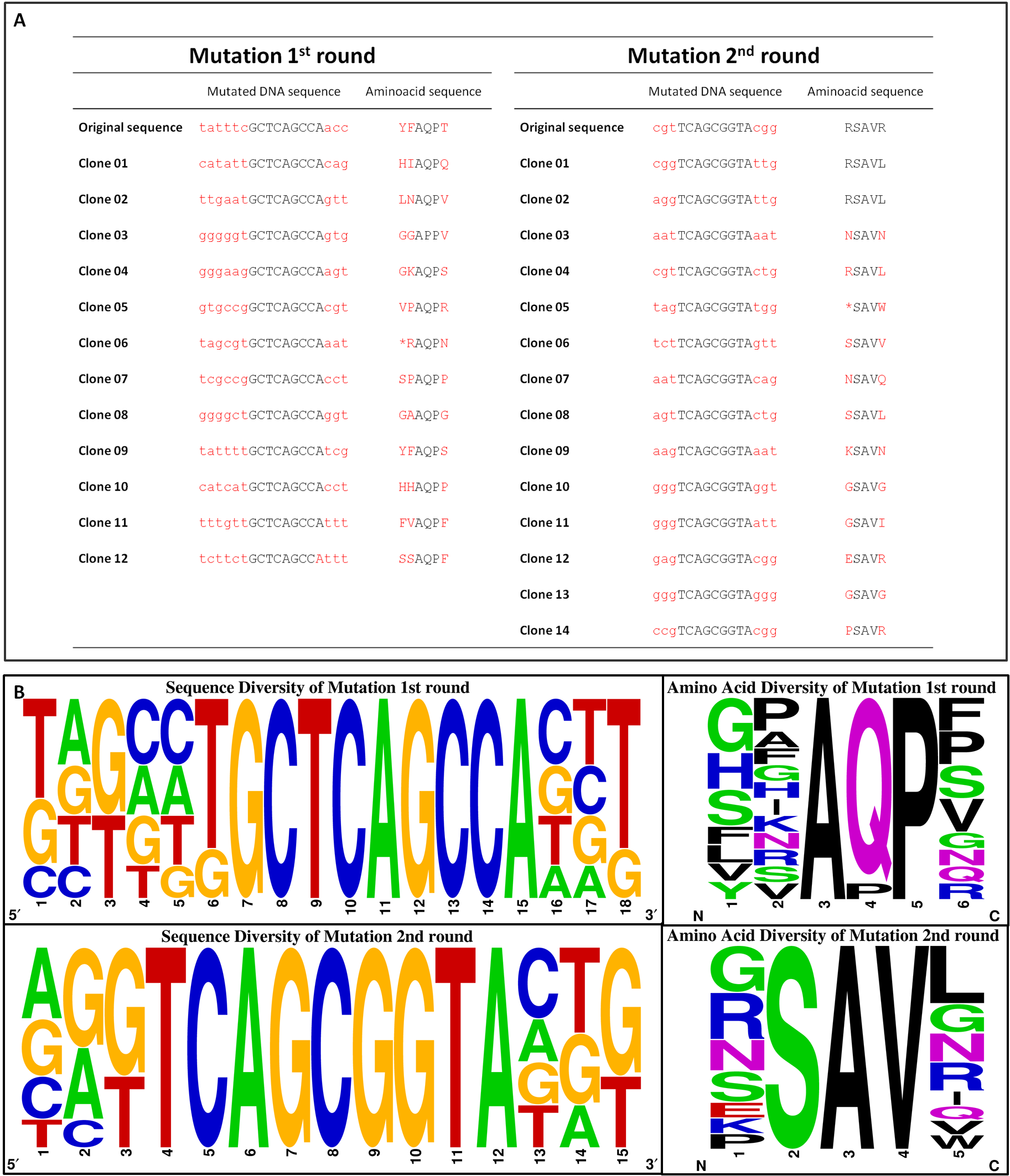
| + | [[File:Gruppe3 Figure04Sequencing diversity02.png|500px|thumb|center| <b>Figure 3: Sequence alignment reflecting the diversity of generated chemoreceptor Tar_QC library. </b> Nucleotide (left) and amino acid (right) sequence alignment of the mutated regions for the <sup>st</sup> round (top) with amino acids mutated at position 149, 150 and 154 and 2<sup>nd</sup> round (bottom) at 69 and 73. Sequencesare represented 5´ to 3´ with increasing numbers standing for the nucleotide or translated amino acid position in the indicated mutation regions, respectively. The consensus of 12 or 14 clones sequenced for 1<sup>st</sup> and 2<sup>nd</sup> round is depicted as a pictogram. The height of each letter represents the frequency of each nucleotide at that position. Original sequences: 1<sup>st</sup> round tatttcGCTCAGCCAacc / YFAQPT; 2<sup>nd</sup> round cgtTCAGCGGTAcgg / RSAVR.G: guanine, C: cytosine, A: adenine, T: thymine.]] | ||
| + | |||
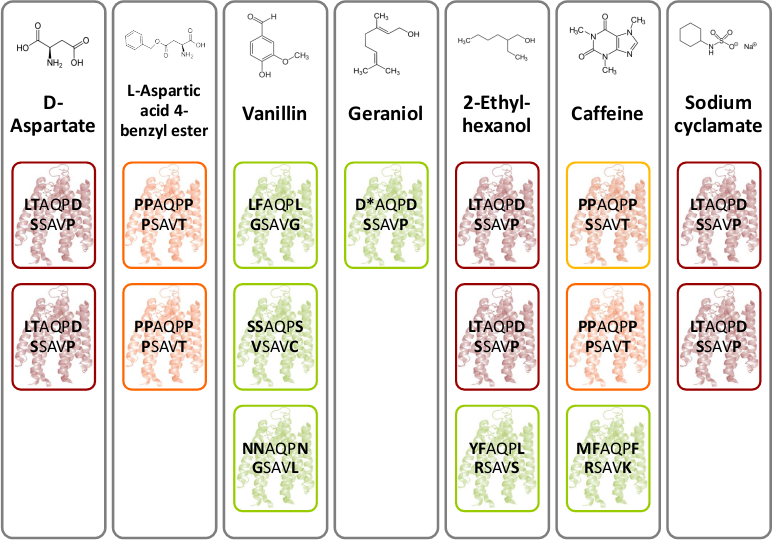
| + | [[File:Gruppe3 Figure03.png|500px|thumb|center| <b>Figure 4: Novel chemoreceptors recognize different tested chemicals. </b> The designed Tar_QC library was transformed into BL21 cells and then tested on swimming agar plates with seven different interesting chemicals. After three rounds of selection of the fastest swimming <i>E. coli</i>s towards these, three clones per chemical were picked. Sequencing results identified two novel chemoreceptors that were found for different chemicals: the novel receptor with amino acid sequence LTAQPD…SSAVP (red boxes) was identified for swimming towards D-Aspartate, Ethylhexanol and Sodium cyclamate and the novel receptor with amino acid residues PAQPP…PSAVT (orange boxes) was identified for swimming towards L-Aspartate and Coffeine. The green boxes show different identified receptors.]] | ||
| + | |||
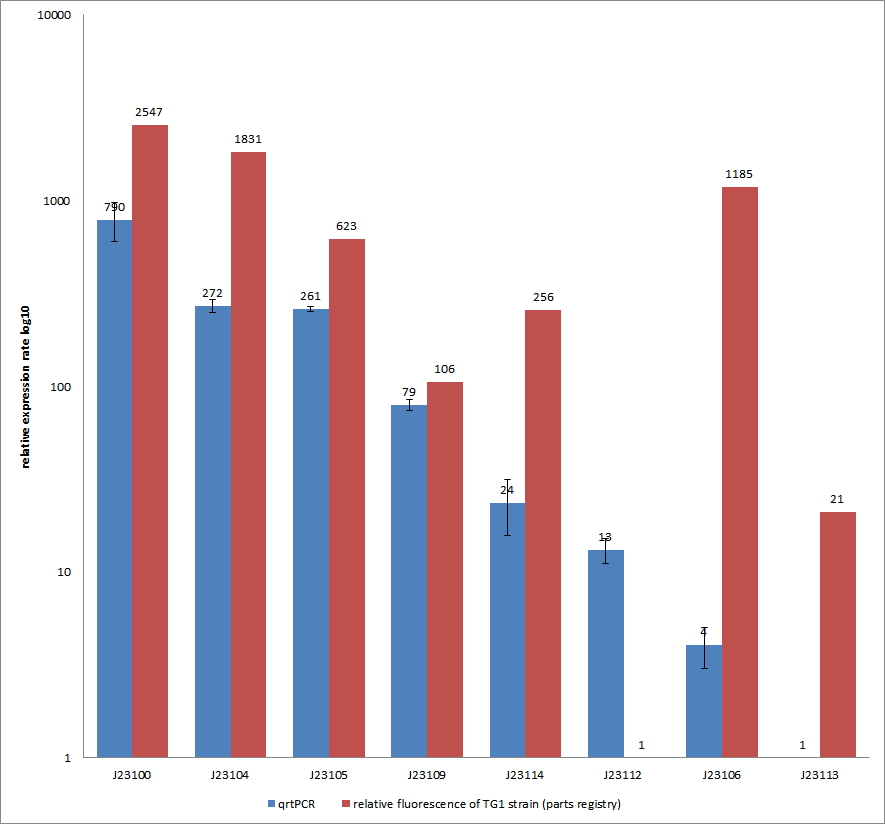
| + | [[File:QrtPCR new.png|500px|thumb|center| <b>Figure 5: Comparison of relative expression rates of constitutive promoters by qrtPCR and relative fluorescence (parts registry).</b> The blue bar indicates the measured expression rates for our constructs (J23100, J23104, J23105, J23109, J23114, J23113, J23106, J23113) and the red ones those for the literature values represented in the “parts registry”. The measurements are illustrated in a logarithmic application. The standard variation was calculated for our measured values (black error bar).]] | ||
To get rid of BioBrick standard restriction sites, the QuikChange reaction is applied. In the case of the Tar receptor, the <i>Bsa</i>I site will be | To get rid of BioBrick standard restriction sites, the QuikChange reaction is applied. In the case of the Tar receptor, the <i>Bsa</i>I site will be | ||
Revision as of 18:56, 25 September 2012
 | |
Deutsch  / English / English  |
 |
Contents |
Homing coli: Engineering E. coli to become tracking dogs
The model organism Escherichia coli is naturally capable of sensing substances in its environment and consequently moves directionally towards these, a phenomenon known as chemotaxis. Here, we apply directed evolution to chemoreceptors by targeting five amino acid residues in the ligand binding site to enable E. coli to perceive novel substances. In order to investigate mobility and directed movement towards a substance, an effective mobility selection method using special "swimming plates" is designed. Additionally, we attempt to improve E. coli's swimming velocity by creating new parts derived from its own motility apparatus. Based on our selection system, we identify variants of chemoreceptors with new binding specificities in the mutant library. By these means, we aim to train the bacterium to detect new molecules such as tumor cell markers. Once having established E. coli as our "tracking dog", the possible applications in medicine but also to environmental issues are virtually countless.
#1 - Selection / Swimming
#2 - Speed Improvement
#3 - Chemoreceptor Library


To get rid of BioBrick standard restriction sites, the QuikChange reaction is applied. In the case of the Tar receptor, the BsaI site will be exchanged while keeping the codon for the same amino acid.
To generate a chemoreceptor library our method of choice requires the absence of the BsaI restriction site in both, the insert and the vector backbone, of the cloned plasmid. Therefore, pUC18 containing one BsaI site in the bla gene seems to be not appropriate. Thus, we moved on cloning our promoter constructs with the quik changed Tar (TAR_QC) at the XbaI restriction site into the pSB1C3 BioBrick vector. There are two advantages coming along: Firstly, we need to send our designed biobricks this year in this particular vector and secondly, this vector contains a chloramphenicol resistance gene and hence, lacking the undesired BsaI site.
| ↑ Back to top! |

|
 "
"