Team:Goettingen/Notebook/Results
From 2012.igem.org
| Line 1: | Line 1: | ||
| - | {{GoettingenHeader}} | + | {{GoettingenHeader|deu=Team:Goettingen/Notebook/Results_deu|eng=Team:Goettingen/Notebook/Results}} |
| - | + | == Homing coli: Engineering <i>E. coli</i> to become tracking dogs == | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
The model organism <i>Escherichia coli</i> is naturally capable of sensing substances in its environment and consequently moves directionally | The model organism <i>Escherichia coli</i> is naturally capable of sensing substances in its environment and consequently moves directionally | ||
towards these, a phenomenon known as chemotaxis. Here, we apply directed evolution to chemoreceptors by targeting five amino acid | towards these, a phenomenon known as chemotaxis. Here, we apply directed evolution to chemoreceptors by targeting five amino acid | ||
| Line 42: | Line 10: | ||
the bacterium to detect new molecules such as tumor cell markers. Once having established <i>E. coli</i> as our "tracking dog", the possible | the bacterium to detect new molecules such as tumor cell markers. Once having established <i>E. coli</i> as our "tracking dog", the possible | ||
applications in medicine but also to environmental issues are virtually countless. | applications in medicine but also to environmental issues are virtually countless. | ||
| - | |||
| - | |||
| + | == #1 - Selection / Swimming == | ||
| + | [[File:Goett_Coli_select.jpg]] | ||
| + | == #2 - Speed Improvement == | ||
| + | [[File:Goett_Coli_speed.jpg]] | ||
| + | == #3 - Chemoreceptor Library == | ||
| + | [[File:Goett_Coli_nose.jpg]] | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
To get rid of BioBrick standard restriction sites, the QuikChange reaction is applied. In the case of the Tar receptor, the <i>Bsa</i>I site will be | To get rid of BioBrick standard restriction sites, the QuikChange reaction is applied. In the case of the Tar receptor, the <i>Bsa</i>I site will be | ||
exchanged while keeping the codon for the same amino acid. | exchanged while keeping the codon for the same amino acid. | ||
| - | + | ||
| - | + | ||
To generate a chemoreceptor library our method of choice requires the absence of the <i>Bsa</i>I restriction site in both, the insert and the vector backbone, | To generate a chemoreceptor library our method of choice requires the absence of the <i>Bsa</i>I restriction site in both, the insert and the vector backbone, | ||
of the cloned plasmid. Therefore, pUC18 containing one <i>Bsa</i>I site in the <i>bla</i> gene seems to be not appropriate. | of the cloned plasmid. Therefore, pUC18 containing one <i>Bsa</i>I site in the <i>bla</i> gene seems to be not appropriate. | ||
| Line 86: | Line 28: | ||
There are two advantages coming along: Firstly, we need to send our designed biobricks this year in this particular vector and secondly, | There are two advantages coming along: Firstly, we need to send our designed biobricks this year in this particular vector and secondly, | ||
this vector contains a chloramphenicol resistance gene and hence, lacking the undesired <i>Bsa</i>I site. | this vector contains a chloramphenicol resistance gene and hence, lacking the undesired <i>Bsa</i>I site. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
{{GoettingenFooter}} | {{GoettingenFooter}} | ||
Revision as of 15:07, 21 September 2012
 | |
Deutsch  / English / English  |
 |
Contents |
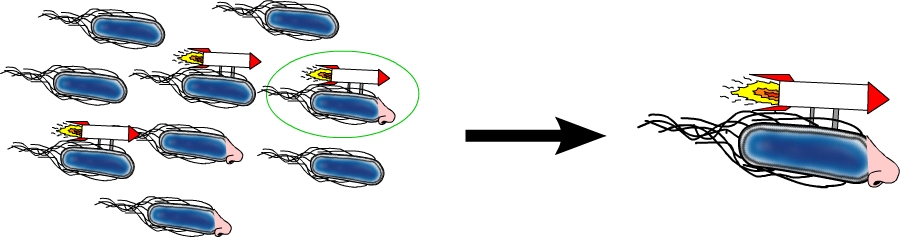
Homing coli: Engineering E. coli to become tracking dogs
The model organism Escherichia coli is naturally capable of sensing substances in its environment and consequently moves directionally towards these, a phenomenon known as chemotaxis. Here, we apply directed evolution to chemoreceptors by targeting five amino acid residues in the ligand binding site to enable E. coli to perceive novel substances. In order to investigate mobility and directed movement towards a substance, an effective mobility selection method using special "swimming plates" is designed. Additionally, we attempt to improve E. coli's swimming velocity by creating new parts derived from its own motility apparatus. Based on our selection system, we identify variants of chemoreceptors with new binding specificities in the mutant library. By these means, we aim to train the bacterium to detect new molecules such as tumor cell markers. Once having established E. coli as our "tracking dog", the possible applications in medicine but also to environmental issues are virtually countless.
#1 - Selection / Swimming
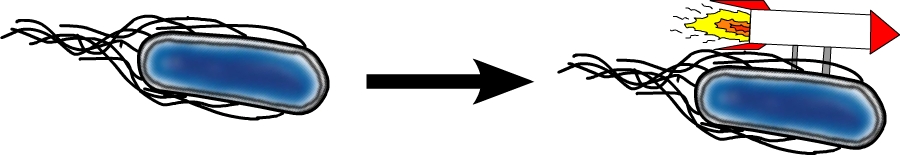
#2 - Speed Improvement
#3 - Chemoreceptor Library
To get rid of BioBrick standard restriction sites, the QuikChange reaction is applied. In the case of the Tar receptor, the BsaI site will be exchanged while keeping the codon for the same amino acid.
To generate a chemoreceptor library our method of choice requires the absence of the BsaI restriction site in both, the insert and the vector backbone, of the cloned plasmid. Therefore, pUC18 containing one BsaI site in the bla gene seems to be not appropriate. Thus, we moved on cloning our promoter constructs with the quik changed Tar (TAR_QC) at the XbaI restriction site into the pSB1C3 BioBrick vector. There are two advantages coming along: Firstly, we need to send our designed biobricks this year in this particular vector and secondly, this vector contains a chloramphenicol resistance gene and hence, lacking the undesired BsaI site.
| ↑ Back to top! |

|
 "
"