Team:University College London/Module 3/Modelling
From 2012.igem.org
Erinoerton (Talk | contribs) (→Modelling) |
Erinoerton (Talk | contribs) (→Modelling) |
||
| Line 15: | Line 15: | ||
| POPex || 0.0 || Persistent organic pollutants (ex = extracellular) that are not adhered to plastic surface | | POPex || 0.0 || Persistent organic pollutants (ex = extracellular) that are not adhered to plastic surface | ||
|- | |- | ||
| - | | PEPOPex || 9.24E-5 || Persistent organic pollutants (ex = extracellular) that are adhered to the plastic surface | + | | PEPOPex || 9.24E-5 || Persistent organic pollutants (ex = extracellular) that are adhered to the plastic surface<sup>3</sup> |
|- | |- | ||
| - | | POPin || 0.5 || Persistent organic pollutants (in = intracellular) assumed from <i>E. coli</i> membrane permeability | + | | POPin || 0.5 || Persistent organic pollutants (in = intracellular) assumed from <i>E. coli</i> membrane permeability <sup>4</sup> |
|- | |- | ||
| mRNANahR || 0.0|| NahR mRNA product | | mRNANahR || 0.0|| NahR mRNA product | ||
| Line 40: | Line 40: | ||
! Number !! Reaction !! Reaction rate (molecules/sec) !! Notes | ! Number !! Reaction !! Reaction rate (molecules/sec) !! Notes | ||
|- | |- | ||
| - | | R1 || PE + POPex ↔ PEPOPex || Forward: 1000 <br /> Backward: 1 || Pops have 1000 to 10000 times greater tendency to adhere to plastic than float free in the ocean | + | | R1 || PE + POPex ↔ PEPOPex || Forward: 1000 <br /> Backward: 1 || Pops have 1000 to 10000 times greater tendency to adhere to plastic than float free in the ocean<sup>5</sup> |
|- | |- | ||
| - | | R2 || POPex ↔ POPin || Forward: 0.6 <br /> Backward: 0.4 || Based on membrane permeability: diffusion gradient | + | | R2 || POPex ↔ POPin || Forward: 0.6 <br /> Backward: 0.4 || Based on membrane permeability<sup>4</sup>: diffusion gradient |
|- | |- | ||
| - | | R3 || POPin + mRNA.Nahr → POPin.mRNA.Nahr || Forward: 1 <br /> Backward: 0.0001 || Based on the assumption that the chemical structure/size of POPs is similar to salycilate. Salycilate binds to the NahR mRNA product, which complex then binds to the pSal promoter. | + | | R3 || POPin + mRNA.Nahr → POPin.mRNA.Nahr || Forward: 1 <br /> Backward: 0.0001 || Based on the assumption that the chemical structure/size of POPs is similar to salycilate<sup>6</sup>. Salycilate binds to the NahR mRNA product, which complex then binds to the pSal promoter. |
|- | |- | ||
| - | | R9 || 0 → mRNA.Nahr || Forward: 0.088 <br /> Backward: 0.6 || Transcription rate of NahR in molecules/sec (for NahR size 909 bp<sup> | + | | R9 || 0 → mRNA.Nahr || Forward: 0.088 <br /> Backward: 0.6 || Transcription rate of NahR in molecules/sec (for NahR size 909 bp<sup>7</sup>, transcription rate in E.coli 80bp/sec<sup>8</sup>) under constitutive promoter control |
|- | |- | ||
| - | | R4 || POPinmRNANahr → POPinmRNANahr.Psal || Forward: 78200 <br /> Backward: 0.191 <sup> | + | | R4 || POPinmRNANahr → POPinmRNANahr.Psal || Forward: 78200 <br /> Backward: 0.191 <sup>9</sup> || NahR to pSal binding based on the assumption that POP-NahR binding has no effect on NahR-pSal binding |
|- | |- | ||
| - | | R5 || POPexmRNANahr.Psal → Lin.mRNA|| 0.054 || Transcription rate of Laccase in molecules/sec (for laccase size 1500 bp<sup> | + | | R5 || POPexmRNANahr.Psal → Lin.mRNA|| 0.054 || Transcription rate of Laccase in molecules/sec (for laccase size 1500 bp<sup>10</sup>, transcription rate in E.coli 80bp/sec<sup>8</sup>) |
|- | |- | ||
| - | | R6 || Lin.mRNA → Lin|| 0.04 || Translation rate of Laccase in molecules/sec (for laccase size 500 aa, translation rate in E.coli 20aa/sec<sup> | + | | R6 || Lin.mRNA → Lin|| 0.04 || Translation rate of Laccase in molecules/sec (for laccase size 500 aa<sup>10</sup>, translation rate in E.coli 20aa/sec<sup>8</sup>) |
|- | |- | ||
| - | | R11 || Lin → Lindegp || 0.03 || Degradation rate of laccase must be taken into account due to suboptimal conditions | + | | R11 || Lin → Lindegp || 0.03 || Degradation rate of laccase<sup>11</sup> must be taken into account due to suboptimal conditions |
|- | |- | ||
| - | | R7 || Lin → Lex || Forward: 0.9 <br /> Backward: 0.1 || Our laccase is a periplasmic enzyme, therefore most of it is released in the periplasm. However, we assumed leakage of 20 percent based on suboptimal conditions in the periplasm<sup> | + | | R7 || Lin → Lex || Forward: 0.9 <br /> Backward: 0.1 || Our laccase is a periplasmic enzyme, therefore most of it is released in the periplasm. However, we assumed leakage of 20 percent based on suboptimal conditions in the periplasm<sup>12</sup>, which is able to degrade polyethene. |
|- | |- | ||
| - | | R8 || Lex → PEdegp || Vm: 0.01 <br /> Km: 0.114 || Michaelis-Menten kinetics is used to represent degradation of polyethylene. As the literature values for the Km and Kcat for the polyethylene by laccase are not yet obtained experimentally for the original starin that was observed being able to degrade polyethylene | + | | R8 || Lex → PEdegp || Vm: 0.01 <br /> Km: 0.114 || Michaelis-Menten kinetics is used to represent degradation of polyethylene. As the literature values for the Km and Kcat for the polyethylene by laccase are not yet obtained experimentally for the original starin that was observed being able to degrade polyethylene<sup>13</sup> we made an assumption that degradation of polyethylene is similar to that of lignin (due to polymeric nature of both), values for both Km and Kcat were used from<sup>14</sup> |
|} | |} | ||
| Line 75: | Line 75: | ||
2. Andrady AL (2011) Microplastics in the marine environment. <i>Marine Pollution Bulletin</i> 62: 1596-1605 | 2. Andrady AL (2011) Microplastics in the marine environment. <i>Marine Pollution Bulletin</i> 62: 1596-1605 | ||
| - | + | 3. To follow | |
| - | + | 4. To follow | |
| - | + | 5. Mato Y, Isobe T, Takada H, Kanehiro H, Ohtake C, Kaminuma T (2001) Plastic Resin Pellets as a Transport Medium for Toxic Chemicals in the Marine Environment. <i>Environ. Sci. Technol.</i> 35: 318-324 | |
| - | + | 6. https://2011.igem.org/Team:Peking_S/project/wire/harvest | |
| - | 7. http:// | + | 7. NahR size: http://www.xbase.ac.uk/genome/azoarcus-sp-bh72/NC_008702/azo2419;nahR1/viewer |
| - | 8. Young K, Silver LL (1991) Leakage of periplasmic enzymes from envA1 strains of Escherichia coli. <i>J Bacteriol.</i> 173: 3609–3614 | + | 8. http://kirschner.med.harvard.edu/files/bionumbers/fundamentalBioNumbersHandout.pdf |
| + | |||
| + | 9. Park H, Lim W, Shin H (2005) In vitro binding of purified NahR regulatory protein with promoter Psal. <i>Biochimica et Biophysica Acta</i> 1775: 247-255 | ||
| + | |||
| + | 10. Laccase size: http://partsregistry.org/Part:BBa_K729002 | ||
| + | |||
| + | 11. To follow | ||
| + | |||
| + | 12. Young K, Silver LL (1991) Leakage of periplasmic enzymes from envA1 strains of Escherichia coli. <i>J Bacteriol.</i> 173: 3609–3614 | ||
| + | |||
| + | 13. To follow (c208 ref??) | ||
| + | |||
| + | 14. 5. Ding Z, Peng L, Chen Y, Zhang L, Gu Z, Shi G, Zhang K (2012) Production and characterization of thermostable laccase from the mushroom, Ganoderma lucidum, using submerged fermentation. <i>African Journal of Microbiology Research</i> 6: 1147-1157. DOI: 10.5897/AJMR11.1257 | ||
| + | |||
| + | |||
| + | |||
| + | ==Misc== | ||
| + | 4. Teuten E, Saquing J, Knappe D et al. (2009) Transport and release of chemicals from plastics to the environment and to wildlife. <i>Philosophical transactions of the Royal Society of London</i> 364: 2027-2045 | ||
11.Van A, Rochman C, Flores E, Hill K, Vargas E, Vargas S, Hoh E (2012) Persistent organic pollutants in plastic marine debris found on beaches in San Diego, California. <i>Chemosphere</i> 86: 258-263 | 11.Van A, Rochman C, Flores E, Hill K, Vargas E, Vargas S, Hoh E (2012) Persistent organic pollutants in plastic marine debris found on beaches in San Diego, California. <i>Chemosphere</i> 86: 258-263 | ||
12. Santo M, Weitsman R, Sivan A (2012) The role of the copper-binding enzyme - laccase - in the biodegradation of polyethylene by the actinomycete <i>Rhodococcus ruber. International Biodeterioration & Biodegradation</i> 208: 1-7 | 12. Santo M, Weitsman R, Sivan A (2012) The role of the copper-binding enzyme - laccase - in the biodegradation of polyethylene by the actinomycete <i>Rhodococcus ruber. International Biodeterioration & Biodegradation</i> 208: 1-7 | ||
| - | |||
| Line 101: | Line 117: | ||
Heskett M, Takada H, Yamashita R, Yuyama M, Ito M, Geok YB, Ogata Y, Kwan C, Heckhausen A, Taylor H, Powell T, Morishige C, Young D, Patterson H, Robertson B, Bailey E, Mermoz J (2012) Measurement of persistent organic pollutants (POPs) in plastic resin pellets from remote islands: Toward establishment of background concentrations for International Pellet Watch. <i>Marine Pollution Bulletin</i> 64: 445-448 | Heskett M, Takada H, Yamashita R, Yuyama M, Ito M, Geok YB, Ogata Y, Kwan C, Heckhausen A, Taylor H, Powell T, Morishige C, Young D, Patterson H, Robertson B, Bailey E, Mermoz J (2012) Measurement of persistent organic pollutants (POPs) in plastic resin pellets from remote islands: Toward establishment of background concentrations for International Pellet Watch. <i>Marine Pollution Bulletin</i> 64: 445-448 | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
6 Kunamneni A, Plou FJ, Ballesteros A, and Alcalde M. Laccases and their applications: A patent review. Departamento de Biocatálisis, Instituto de Catálisis y Petroleoquímica, CSIC, Cantoblanco, 28049 Madrid, Spain. | 6 Kunamneni A, Plou FJ, Ballesteros A, and Alcalde M. Laccases and their applications: A patent review. Departamento de Biocatálisis, Instituto de Catálisis y Petroleoquímica, CSIC, Cantoblanco, 28049 Madrid, Spain. | ||
Revision as of 12:19, 21 September 2012
Module 3: Degradation
Description | Design | Construction | Characterisation | Modelling | Results | Conclusions
Modelling
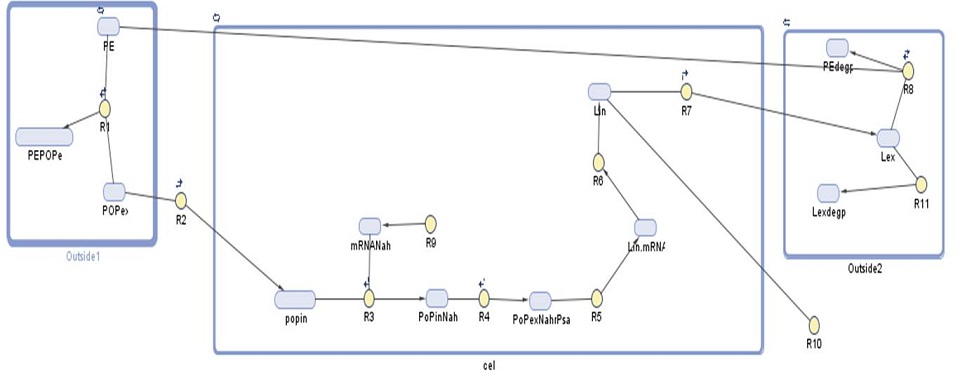
Our gene network model for this module shows the amount of laccase produced by our bacteria. We have used this prediction to find out how many bacteria would be required per cubic meter of sea water in order to effectively degrade polyethylene-based plastics.
| Species | Initial value (molecules) | Notes |
|---|---|---|
| PE | 0.044 | Polyethylene found in North Pacific Gyre per cubic metre1,2 |
| POPex | 0.0 | Persistent organic pollutants (ex = extracellular) that are not adhered to plastic surface |
| PEPOPex | 9.24E-5 | Persistent organic pollutants (ex = extracellular) that are adhered to the plastic surface3 |
| POPin | 0.5 | Persistent organic pollutants (in = intracellular) assumed from E. coli membrane permeability 4 |
| mRNANahR | 0.0 | NahR mRNA product |
| POPinNahR | 0.0 | Complex of the above two molecules |
| POPinNahRpSal | 0.0 | Complex of the above molecule and pSal (promoter that induces laccase transcription) |
| Lin | 0.0 | Intracellular laccase |
| Lex | 0.0 | Extracellular laccase |
| LinmRNA | 0.0 | Laccase mRNA product |
| Ldegp | 0.0 | Laccase that degrades due to suboptimal conditions and malformed laccase that cannot carry out polyethylene degradation |
| PEdegp | 0.0 | Polyethylene degraded by laccase |
| Number | Reaction | Reaction rate (molecules/sec) | Notes |
|---|---|---|---|
| R1 | PE + POPex ↔ PEPOPex | Forward: 1000 Backward: 1 | Pops have 1000 to 10000 times greater tendency to adhere to plastic than float free in the ocean5 |
| R2 | POPex ↔ POPin | Forward: 0.6 Backward: 0.4 | Based on membrane permeability4: diffusion gradient |
| R3 | POPin + mRNA.Nahr → POPin.mRNA.Nahr | Forward: 1 Backward: 0.0001 | Based on the assumption that the chemical structure/size of POPs is similar to salycilate6. Salycilate binds to the NahR mRNA product, which complex then binds to the pSal promoter. |
| R9 | 0 → mRNA.Nahr | Forward: 0.088 Backward: 0.6 | Transcription rate of NahR in molecules/sec (for NahR size 909 bp7, transcription rate in E.coli 80bp/sec8) under constitutive promoter control |
| R4 | POPinmRNANahr → POPinmRNANahr.Psal | Forward: 78200 Backward: 0.191 9 | NahR to pSal binding based on the assumption that POP-NahR binding has no effect on NahR-pSal binding |
| R5 | POPexmRNANahr.Psal → Lin.mRNA | 0.054 | Transcription rate of Laccase in molecules/sec (for laccase size 1500 bp10, transcription rate in E.coli 80bp/sec8) |
| R6 | Lin.mRNA → Lin | 0.04 | Translation rate of Laccase in molecules/sec (for laccase size 500 aa10, translation rate in E.coli 20aa/sec8) |
| R11 | Lin → Lindegp | 0.03 | Degradation rate of laccase11 must be taken into account due to suboptimal conditions |
| R7 | Lin → Lex | Forward: 0.9 Backward: 0.1 | Our laccase is a periplasmic enzyme, therefore most of it is released in the periplasm. However, we assumed leakage of 20 percent based on suboptimal conditions in the periplasm12, which is able to degrade polyethene. |
| R8 | Lex → PEdegp | Vm: 0.01 Km: 0.114 | Michaelis-Menten kinetics is used to represent degradation of polyethylene. As the literature values for the Km and Kcat for the polyethylene by laccase are not yet obtained experimentally for the original starin that was observed being able to degrade polyethylene13 we made an assumption that degradation of polyethylene is similar to that of lignin (due to polymeric nature of both), values for both Km and Kcat were used from14 |
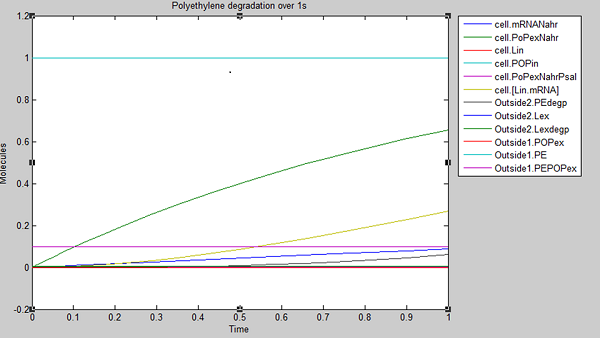
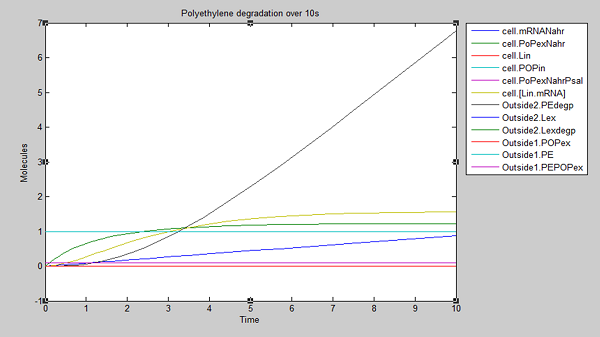
We ran three simulations in SimBiology, each over a different timespan:
1. Goldstein M, Rosenberg M, Cheng L (2012) Increased oceanic microplastic debris enhances oviposition in an endemic pelagic insect, Biology Letters 10.1098
2. Andrady AL (2011) Microplastics in the marine environment. Marine Pollution Bulletin 62: 1596-1605
3. To follow
4. To follow
5. Mato Y, Isobe T, Takada H, Kanehiro H, Ohtake C, Kaminuma T (2001) Plastic Resin Pellets as a Transport Medium for Toxic Chemicals in the Marine Environment. Environ. Sci. Technol. 35: 318-324
6. https://2011.igem.org/Team:Peking_S/project/wire/harvest
7. NahR size: http://www.xbase.ac.uk/genome/azoarcus-sp-bh72/NC_008702/azo2419;nahR1/viewer
8. http://kirschner.med.harvard.edu/files/bionumbers/fundamentalBioNumbersHandout.pdf
9. Park H, Lim W, Shin H (2005) In vitro binding of purified NahR regulatory protein with promoter Psal. Biochimica et Biophysica Acta 1775: 247-255
10. Laccase size: http://partsregistry.org/Part:BBa_K729002
11. To follow
12. Young K, Silver LL (1991) Leakage of periplasmic enzymes from envA1 strains of Escherichia coli. J Bacteriol. 173: 3609–3614
13. To follow (c208 ref??)
14. 5. Ding Z, Peng L, Chen Y, Zhang L, Gu Z, Shi G, Zhang K (2012) Production and characterization of thermostable laccase from the mushroom, Ganoderma lucidum, using submerged fermentation. African Journal of Microbiology Research 6: 1147-1157. DOI: 10.5897/AJMR11.1257
Misc
4. Teuten E, Saquing J, Knappe D et al. (2009) Transport and release of chemicals from plastics to the environment and to wildlife. Philosophical transactions of the Royal Society of London 364: 2027-2045
11.Van A, Rochman C, Flores E, Hill K, Vargas E, Vargas S, Hoh E (2012) Persistent organic pollutants in plastic marine debris found on beaches in San Diego, California. Chemosphere 86: 258-263
12. Santo M, Weitsman R, Sivan A (2012) The role of the copper-binding enzyme - laccase - in the biodegradation of polyethylene by the actinomycete Rhodococcus ruber. International Biodeterioration & Biodegradation 208: 1-7
POPS & PLASTIC QUANTITITES
Rios L, Moore C, Jones P (2007) Persistent organic pollutants carried by synthetic polymers in the ocean environment. Marine Pollution Bulletin 54: 1230–1237
Takada H, et al (2010) Global distribution of organic micropollutants in marine plastics. Report to Algalita Marine Research Institute
Heskett M, Takada H, Yamashita R, Yuyama M, Ito M, Geok YB, Ogata Y, Kwan C, Heckhausen A, Taylor H, Powell T, Morishige C, Young D, Patterson H, Robertson B, Bailey E, Mermoz J (2012) Measurement of persistent organic pollutants (POPs) in plastic resin pellets from remote islands: Toward establishment of background concentrations for International Pellet Watch. Marine Pollution Bulletin 64: 445-448
6 Kunamneni A, Plou FJ, Ballesteros A, and Alcalde M. Laccases and their applications: A patent review. Departamento de Biocatálisis, Instituto de Catálisis y Petroleoquímica, CSIC, Cantoblanco, 28049 Madrid, Spain.
7. BRENDA: EC 1.10.3.2 – laccase
8. E.COLI MEMBRANE PERMEABILITY
Heskett M, Takada H, Yamashita R, Yuyama M, Ito M, Geok YB, Ogata Y, Kwan C, Heckhausen A, Taylor H, Powell T, Morishige C, Young D, Patterson H, Robertson B, Bailey E, Mermoz J (2012) Measurement of persistent organic pollutants (POPs) in plastic resin pellets from remote islands: Toward establishment of background concentrations for International Pellet Watch. Marine Pollution Bulletin 64: 445-448 is this right? This is what’s on excel
7. DIFFERENT REGULATOR SYSTEMS Wang B, Papamichail D, Mueller S, Skiena S (2007) Two Proteins for the Price of One: The Design of Maximally Compressed Coding Sequences. DOI: 10.1.1.174.6416
 "
"