Team:University College London/Module 3/Modelling
From 2012.igem.org
(Difference between revisions)
Erinoerton (Talk | contribs) (→Modelling) |
Erinoerton (Talk | contribs) (→Modelling) |
||
| Line 48: | Line 48: | ||
| PEPOPex || Example | | PEPOPex || Example | ||
|- | |- | ||
| - | | POPin || Dependent on the concentration gradient: when the conc. in is equal to conc. out rate decreases | + | | POPin || Example || Dependent on the concentration gradient: when the conc. in is equal to conc. out rate decreases |
|- | |- | ||
| - | | Nahr || Produced all the time, transcription depends on | + | | Nahr || Example || Produced all the time, transcription depends on |
|- | |- | ||
| - | | POPinNahR || Hill function | + | | POPinNahR || Example || Hill function |
|- | |- | ||
| Lin|| Example | | Lin|| Example | ||
Revision as of 14:46, 20 September 2012
Module 3: Degradation
Description | Design | Construction | Characterisation | Modelling | Results | Conclusions
Modelling
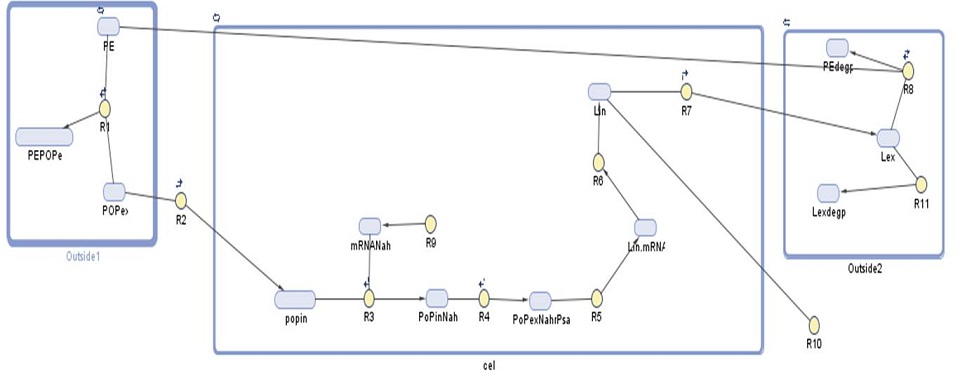
Our gene network model for this module shows the amount of laccase produced by our bacteria. We have used this prediction to find out how many bacteria would be required per cubic meter of sea water in order to effectively degrade polyethylene-based plastics.
| Parameter | Value |
|---|---|
| f.PE+POPex-PEPOPex | Example |
| r.PE+POPEX-PEPOPex | Example |
| f.POPex-POPin | Example |
| r.POPex-POPin | Example |
| f.NahR+POPin-POPinNahR | Example |
| r.NahR+POPin-POPinNahr | Example |
| POPinNahR-Lin+POPinNahR | Example |
| Lin-Lex | Example |
| Lin-Lex | Example |
| f.Null-NahR | Example |
| r.Null-NahR | Example |
| PEPOP+Lex-Lex | Example |
| Species | Initial value | Notes |
|---|---|---|
| PE | Example | |
| POPEX | Example | |
| Lex | Example | |
| PEPOPex | Example | |
| POPin | Example | Dependent on the concentration gradient: when the conc. in is equal to conc. out rate decreases |
| Nahr | Example | Produced all the time, transcription depends on |
| POPinNahR | Example | Hill function |
| Lin | Example |
 "
"