Team:University College London/LabBook/Week6
From 2012.igem.org
Rwilkinson (Talk | contribs) (→Friday 20.7.12) |
|||
| Line 2: | Line 2: | ||
</html>{{:Team:University_College_London/templates/labbookmenu}}<html> | </html>{{:Team:University_College_London/templates/labbookmenu}}<html> | ||
<img src="https://static.igem.org/mediawiki/2012/d/d4/Ucl2012-labbook-monfri.png" /> | <img src="https://static.igem.org/mediawiki/2012/d/d4/Ucl2012-labbook-monfri.png" /> | ||
| - | <img src="https://static.igem.org/mediawiki/2012/e/ec/Ucl2012-labbook-graph6-1.png" /> | + | <img id="6-1" src="https://static.igem.org/mediawiki/2012/e/ec/Ucl2012-labbook-graph6-1.png" /> |
<div class="experimentContent"></html> | <div class="experimentContent"></html> | ||
== Tuesday 17.7.12 == | == Tuesday 17.7.12 == | ||
| Line 54: | Line 54: | ||
<html></div> | <html></div> | ||
<div class="experiment"></div> | <div class="experiment"></div> | ||
| - | <img src="https://static.igem.org/mediawiki/2012/5/58/Ucl2012-labbook-graph6-2.png" /><div class="experimentContent"> | + | <img id="6-2" src="https://static.igem.org/mediawiki/2012/5/58/Ucl2012-labbook-graph6-2.png" /><div class="experimentContent"> |
</html> | </html> | ||
| Line 116: | Line 116: | ||
<html> | <html> | ||
</div><div class="experiment"></div> | </div><div class="experiment"></div> | ||
| - | <img src="https://static.igem.org/mediawiki/2012/8/8e/Ucl2012-labbook-graph6-3.png" /><div class="experimentContent"> | + | <img id="6-3" src="https://static.igem.org/mediawiki/2012/8/8e/Ucl2012-labbook-graph6-3.png" /><div class="experimentContent"> |
</html> | </html> | ||
Revision as of 13:20, 2 August 2012



Contents |
Tuesday 17.7.12
Aim -Testing the competency of the Reinvigorated Cell Line (W3110): After our original attempt to generate competency of the W3100 cell line was inadequate, we undertook a reinvigoration of the W3100 cell line (Expt 5.2). Here we aim to test the competency of the reinvigorated cell line using the same protocol as used in Expt 5.3, by evaluating their growth after transformation with a plasmid of a known high concentration (297ng/ul).
Method
(LOGO) Transformation Protocol 2
Step 1 – Thawing Cells: Use the reinvigorated W3100 cell line created in Week 5 (Expt 5.2)
Step 3 – Addition of BioBrick: To one 2ml eppendorf, add 1ul of pTop plasmid, and to another add nothing – this will be a control.
Step 7 – Adding Broth: SOC media was used, as it is preferred for this protocol.
Step 8 - Incubation: The table below indicates the Ampicillin concentration of the Agar gels.
| Samples | Volume Innoculated | Antibiotic in Gel (conc) | |
|---|---|---|---|
| Plasmid | pTOP | 36ul | Ampicillin (50ug/ml) |
| Control | Positive (No Plasmid) | 36ul | No Antibiotic |
| Negative (No Plasid) | 36ul | Ampicillin (50ug/ml) | |
Wednesday (18.7.12)
Aim - Results from Transformation
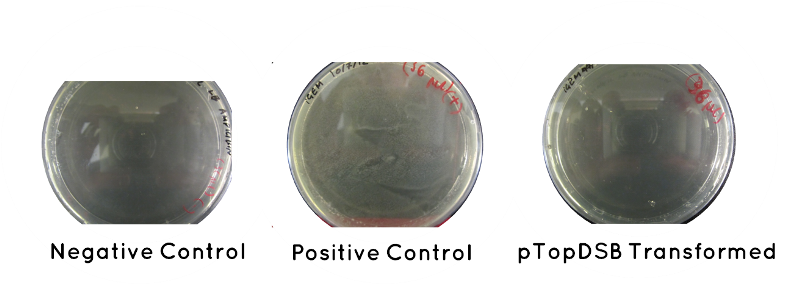
Result: Table below indicates there was no growth for our cells, but that the controls worked. Also included is an image of each plate.
| Samples | Growth/No Growth | |
|---|---|---|
| Plasmid | pTOP | No Growth |
| Control | Positive (No Plasmid) | Growth |
| Negative (No Plasid) | No Growth | |

Wednesday (18.7.12)
Aim - Testing the transformation protocols: In light of the problems with cell competency, we decided to test whether the choice of protocol was contributing to the poor transformation results – especially as it would require another week to set up another cell line. We used a known competent cell line, and tested its competency against both of our cell lines, for Transformation Protocol 1 vs Transformation Protocol 2. Each sample was plated on an Ampicillin positive Agar and incubated overnight.
(LOGO) Transformation - Protocol 1
(LOGO) Transformation - Protocol 2
Step ?: The table below describes the plates necessary for this investigation, each of which shoud carry Ampicillin antibiotic at a concentration of 50ug/ml.
| Plates | |
|---|---|
| Protocol 1 | W3100 – Original |
| W3100 – Reinvigorated | |
| W3100 – Known Competent | |
| Protocol 2 | W3100 – Original |
| W3100 – Reinvigorated | |
| W3100 – Known Competent | |
Thursday 19.7.12
Aim - Results from Transformation of pTop
Results: Table below indicates there was significant growth from the Original W3100 cells, but not from other cell lines.
| Plates | Colony Formation | |
|---|---|---|
| Protocol 1 | W3100 – Original | Yes |
| W3100 – Reinvigorated | No | |
| W3100 – Known Competent | No | |
| Protocol 2 | W3100 – Original | Yes |
| W3100 – Reinvigorated | No | |
| W3100 – Known Competent | No | |
Conclusion: The Original Cell line appears to be competent for both Protocols, while our reinvigorated cell line does not appear to be competent with either. The cells known to be competent showed no growth, for reasons that are unclear. For our Original W3100 cell line, there was greater colony formation with Protocol 1 than with Protocol 2, so we shall proceed with this protocol. It is likely that the failure of our transformations is due to the switch to protocol 2, and the use of too little BioBrick to transform our cells. The protocol recommends 1-2ul, but members around the lab generally use 5ul for transforming cells. We will take this approach and see if our transformations improve.

Thursday 19.7.12
Aim – Transformations: Expt 6.3 will be transforming our main Constitutive Promoter - BBa_J23119 – which is to be used in a number of modules. It will also aim to transform the Gas Vesicle Polycistronic Gene BBa_I750016, which is required for the production of gas vesicles. Finally, it will also transform the Ribosome Binding Site BBa_B0034, which failed to transform in Expt 5.1, and the Double Termination BBa_B0015. Both of these are key to all modules. Method:
(LOGO) Transformation Protocol 1
Step 1 – Thawing Cells: Use W3100 cell line created in Week 2 (Expt 2.1)
Step 3 – Addition of BioBrick: To each 2ml eppendorf, add 1ul of the following BioBricks. Note: we have changed the protocol for our positive control. Previously it contained no BioBrick, but it has been recommended to us that we transform our positive control such that there is one for each BioBrick – this will tell us if the BioBrick has in any way affected cell viability. This will be used from this point onwards. Include an extra tube as a negative control, with no BioBrick added
| Function | Module | ||
|---|---|---|---|
| BioBrick | BBa_J23119 | Constitutive Promoter | Degradation/
Salt Tolerance/ Containment |
| BBa_I750016 | Gas Vesicle Polycistronic Gene | Buoyancy | |
| BBa_B0015 | Double Terminator | All | |
| BBa_B0034 | Ribosome Binding Site (RBS) | All | |
| Control | (No BioBricks) | ||
Step 9 – Plating samples on Agar Plates: The table below indicates the chosen inoculation volume (two for each BioBrick) and the correct gel antibiotic concentration for all samples.
| Samples | Volume Inoculated | Antibiotic in Gel (ug/ml) | |
|---|---|---|---|
| BioBrick | BBa_J23119 | 10ul | Ampicillin(50ug/ml) |
| 90ul | |||
| BBa_I750016 | 10ul | ||
| 90ul | |||
| BBa_B0015 | 10ul | ||
| 90ul | |||
| BBa_B0034 | 10ul | ||
| 90ul | |||
| Control | Positive Positive (Contains BioBrick – one for each of the above) | 36ul | No Antibiotic |
| Negative (No BioBrick) | 36ul | 1x Ampicillin(50ug/ml) | |
Friday 20.7.12
Aim 1 - Results of Transformation Result: Table below indicates there was growth for all of our transformed cells, and that the controls worked. Also included is an image of each plate.
| Samples | Volume Inoculated | Growth/ No Growth | |
|---|---|---|---|
| BioBrick | BBa_J23119 | 10ul | Growth |
| 90ul | Growth | ||
| BBa_I750016 | 10ul | Growth | |
| 90ul | Growth | ||
| BBa_B0015 | 10ul | Growth | |
| 90ul | Growth | ||
| BBa_B0034 | 10ul | Growth | |
| 90ul | Growth | ||
| Control | Positive Positive (Contains BioBrick – one for each of the above) | 36ul | Growth |
| Negative (No BioBrick) | 36ul | No Growth | |
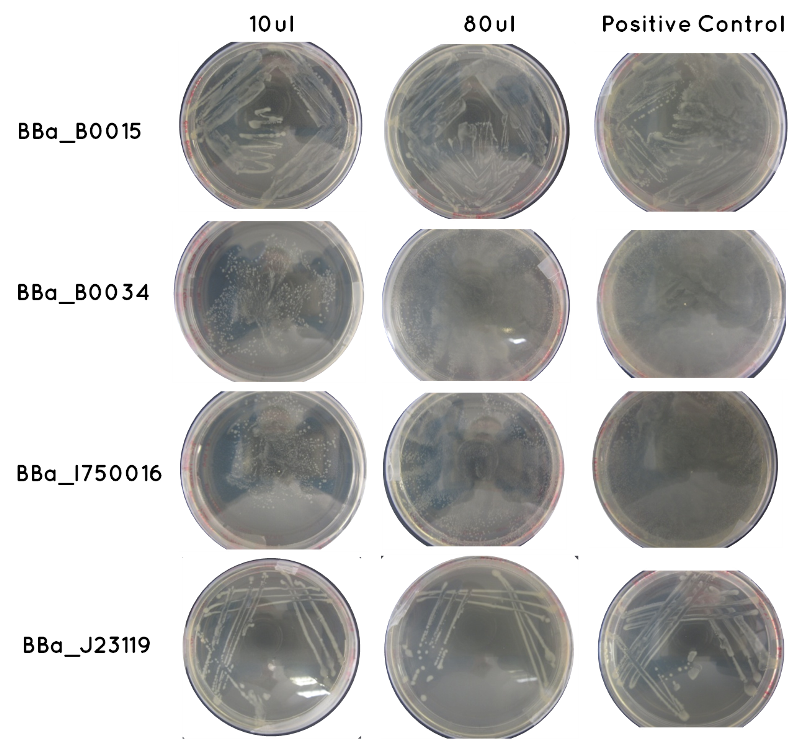
Conclusion: As of week 7 we can continue with colony picking for these agar plates, which will enable us to culture our transformed cells, and purify the plasmid. The purified plasmid will undergo restriction digest to determine the presence of the correct BioBrick inserts.
 "
"