Team:UNAM Genomics Mexico/Modeling/Cross Talk
From 2012.igem.org
| Line 138: | Line 138: | ||
<br /> | <br /> | ||
| + | '''For the intrinsic TF XylR in Bacillus subtilis compared against the promoter Pbad:''' | ||
| + | |||
| + | Matrices | ||
| + | matrix name ncol nrow pseudo Wmin Wmax Wrange | ||
| + | 1 matrix-scan_2012-09-23 23 6 0 -66.200 28.700 94.900 a:0.291 c:0.207 g:0.204 t:0.297 | ||
| + | Number of sequences scanned 0 | ||
| + | Sum of sequence lengths 0 | ||
| + | N residues 0 | ||
| + | Matches per matrix | ||
| + | matrix name matches scored | ||
| + | 1 matrix-scan_2012-09-23 0 0 | ||
| + | TOTAL 0 0 | ||
| + | Host name miztli.nnb.unam.mx | ||
| + | Job started 2012-09-23.134705 | ||
| + | Job done 2012-09-23.134705 | ||
| + | Seconds 0.13 | ||
| + | user 0.13 | ||
| + | system 0.03 | ||
| + | cuser 0.1 | ||
| + | csystem 0.02 | ||
| + | |||
| + | Matrix logo: | ||
| + | |||
| + | [[File:Unamgenomicsmodellogo4.jpg]] | ||
}} | }} | ||
Revision as of 22:15, 26 September 2012

Cross Talk
| abi Nanotubes |
abi1 The logic |
Random info |
Due to the fact that our construction introduces exogenous transcription factors to the cell, we wanted to make sure that there would be no crosstalk, meaning that they wouldn’t recognize other promoters and create noise in the system. For this purpose, we downloaded weight matrixes for the sequences recognized by our transcription factors from PRODORIC database, and we downloaded the sequences for promoters (-200, +50 parting from the TSS) using retrieve sequence from RSA-tools. We chose Bacillus subtilis strain 168 because our strain is a derivative of this strain (I like the word strain…strain, strain, strain yay!). After that, we used quick matrix-scan in RSA-tools (Regulation Sequence Analysis-tools) to compare the sequences of the promoters to the weight matrixes to see if any of our transcription factors could bind in a troublesome place. We soon found out that this was not the case. Satisfied with our new-found self-confidence, we proceeded undauntedly to explore the more obscure parts of our project as our doubts wore away in the computational demonstration that they were, in fact, unworthy of being called truths.
Here are the boring details of the deed:
Details summary:
Tf |
Sequence |
Matches |
AraC belonging to E. coli |
Bacillus subtilis promoters |
0 |
AraR belonging to B. subtilis |
Pbad/Pxyl promoter |
0 |
LasR belonging to P. aeruginosa |
Bacillus subtilis promoters |
0 |
XylR belonging to B. subtilits |
Pbad promoter |
0 |
XylR belonging to E. coli |
Bacillus subtilis promoters |
0 |
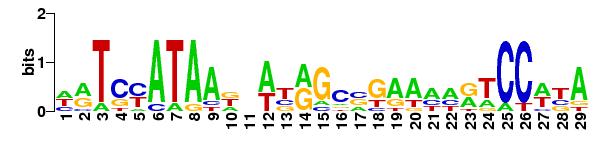
For AraC compared against the promoters in Bacillus subtilis strain 168:
Matrices
matrix name ncol nrow pseudo Wmin Wmax Wrange
1 matrix-scan_2012-09-23 33 6 0 -101.700 44.600 146.300 a:0.327 c:0.165 g:0.193 t:0.314
Number of sequences scanned 4176
Sum of sequence lengths 498968
N residues 14
Matches per matrix
matrix name matches scored
1 matrix-scan_2012-09-23 0 0
TOTAL 0 0
Host name miztli.nnb.unam.mx
Job started 2012-09-23.134423
Job done 2012-09-23.134424
Seconds 0.39
user 0.39
system 0.02
cuser 0.09
csystem 0.03
The logo of the matrix used:
For AraR compared against the promoter Pbad:
Matrices
matrix name ncol nrow pseudo Wmin Wmax Wrange
1 matrix-scan_2012-09-23 16 6 0 -47.000 16.000 63.000 a:0.291 c:0.207 g:0.204 t:0.297
Number of sequences scanned 0
Sum of sequence lengths 0
N residues 0
Matches per matrix
matrix name matches scored
1 matrix-scan_2012-09-23 0 0
TOTAL 0 0
Host name miztli.nnb.unam.mx
Job started 2012-09-23.135941
Job done 2012-09-23.135941
Seconds 0.14
user 0.14
system 0.02
cuser 0.1
csystem 0.02
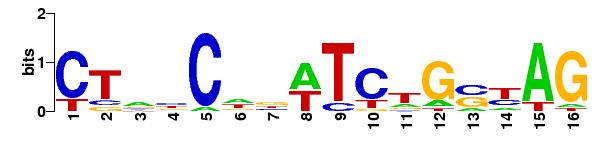
For LasR compared against the promoters in Bacillus subtilis strain 168:
Matrices
matrix name ncol nrow pseudo Wmin Wmax Wrange
1 matrix-scan_2012-09-23 16 6 0 -47.500 13.500 61.000 a:0.327 c:0.165 g:0.193 t:0.314
Number of sequences scanned 4176
Sum of sequence lengths 498968
N residues 14
Matches per matrix
matrix name matches scored
1 matrix-scan_2012-09-23 0 0
TOTAL 0 0
Host name miztli.nnb.unam.mx
Job started 2012-09-23.140435
Job done 2012-09-23.140437
Seconds 0.4
user 0.4
system 0.02
cuser 0.09
csystem 0.03
The logo of the matrix:
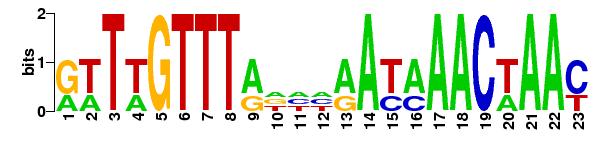
For the intrinsic TF XylR in Bacillus subtilis compared against the promoter Pbad:
Matrices matrix name ncol nrow pseudo Wmin Wmax Wrange 1 matrix-scan_2012-09-23 23 6 0 -66.200 28.700 94.900 a:0.291 c:0.207 g:0.204 t:0.297 Number of sequences scanned 0 Sum of sequence lengths 0 N residues 0 Matches per matrix matrix name matches scored 1 matrix-scan_2012-09-23 0 0 TOTAL 0 0 Host name miztli.nnb.unam.mx Job started 2012-09-23.134705 Job done 2012-09-23.134705 Seconds 0.13 user 0.13 system 0.03 cuser 0.1 csystem 0.02
Matrix logo:
 "
"