Team:University College London/LabBook/Week6
From 2012.igem.org
Rwilkinson (Talk | contribs) (→Friday 20.7.12) |
Sednanalien (Talk | contribs) (→Friday 20.7.12) |
||
| (13 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
</html>{{:Team:University_College_London/templates/labbookmenu}}<html> | </html>{{:Team:University_College_London/templates/labbookmenu}}<html> | ||
<img src="https://static.igem.org/mediawiki/2012/d/d4/Ucl2012-labbook-monfri.png" /> | <img src="https://static.igem.org/mediawiki/2012/d/d4/Ucl2012-labbook-monfri.png" /> | ||
| - | <img src="https://static.igem.org/mediawiki/2012/e/ec/Ucl2012-labbook-graph6-1.png" /> | + | <img id="6-1" src="https://static.igem.org/mediawiki/2012/e/ec/Ucl2012-labbook-graph6-1.png" /> |
<div class="experimentContent"></html> | <div class="experimentContent"></html> | ||
== Tuesday 17.7.12 == | == Tuesday 17.7.12 == | ||
| - | '''Aim -Testing the competency of the Reinvigorated Cell Line (W3110):''' After our original attempt to generate competency of the | + | '''Aim -Testing the competency of the Reinvigorated Cell Line (W3110):''' After our original attempt to generate competency of the W3110 cell line was inadequate, we undertook a reinvigoration of the W310 cell line (Expt 5.2). Here we aim to test the competency of the reinvigorated cell line using the same protocol as used in Expt 5.3, by evaluating their growth after transformation with a plasmid of a known high concentration (297ng/ul). |
'''Method''' | '''Method''' | ||
| - | + | <html><div class="protocol protocol-Transformation">Transformation Protocol 2</div><div class="protocolContent"></html>{{:Team:University_College_London/Protocols/Transformation2}}<html></div></html> | |
| - | Step 1 – Thawing Cells: Use the reinvigorated | + | Step 1 – Thawing Cells: Use the reinvigorated W3110 cell line created in Week 5 (Expt 5.2) |
Step 3 – Addition of BioBrick: To one 2ml eppendorf, add 1ul of pTop plasmid, and to another add nothing – this will be a control. | Step 3 – Addition of BioBrick: To one 2ml eppendorf, add 1ul of pTop plasmid, and to another add nothing – this will be a control. | ||
| Line 50: | Line 50: | ||
|} | |} | ||
| + | {{:Team:University_College_London/templates/pictureinsert|title=Picture 1|url=images/3/36/UCLiGEM2012Agar_6.1.2.png}} | ||
<html></div> | <html></div> | ||
<div class="experiment"></div> | <div class="experiment"></div> | ||
| - | <img src="https://static.igem.org/mediawiki/2012/5/58/Ucl2012-labbook-graph6-2.png" /><div class="experimentContent"> | + | <img id="6-2" src="https://static.igem.org/mediawiki/2012/5/58/Ucl2012-labbook-graph6-2.png" /><div class="experimentContent"> |
</html> | </html> | ||
| Line 60: | Line 61: | ||
'''Aim - Testing the transformation protocols:''' In light of the problems with cell competency, we decided to test whether the choice of protocol was contributing to the poor transformation results – especially as it would require another week to set up another cell line. We used a known competent cell line, and tested its competency against both of our cell lines, for Transformation Protocol 1 vs Transformation Protocol 2. Each sample was plated on an Ampicillin positive Agar and incubated overnight. | '''Aim - Testing the transformation protocols:''' In light of the problems with cell competency, we decided to test whether the choice of protocol was contributing to the poor transformation results – especially as it would require another week to set up another cell line. We used a known competent cell line, and tested its competency against both of our cell lines, for Transformation Protocol 1 vs Transformation Protocol 2. Each sample was plated on an Ampicillin positive Agar and incubated overnight. | ||
| - | + | <html><div class="protocol protocol-Transformation">Transformation Protocol 1</div><div class="protocolContent"></html>{{:Team:University_College_London/Protocols/Transformation1}}<html></div></html> | |
| - | + | <html><div class="protocol protocol-Transformation">Transformation Protocol 2</div><div class="protocolContent"></html>{{:Team:University_College_London/Protocols/Transformation2}}<html></div></html> | |
'''Step ?:''' The table below describes the plates necessary for this investigation, each of which shoud carry Ampicillin antibiotic at a concentration of 50ug/ml. | '''Step ?:''' The table below describes the plates necessary for this investigation, each of which shoud carry Ampicillin antibiotic at a concentration of 50ug/ml. | ||
| Line 70: | Line 71: | ||
! colspan="2" |Plates | ! colspan="2" |Plates | ||
|- | |- | ||
| - | | rowspan="3" | Protocol 1 || | + | | rowspan="3" | Protocol 1 || W3110 – Original |
|- | |- | ||
| - | | | + | | W3110 – Reinvigorated |
|- | |- | ||
| - | | | + | | W3110 – Known Competent |
|- | |- | ||
| - | | rowspan="3" | Protocol 2 || | + | | rowspan="3" | Protocol 2 || W3110 – Original |
|- | |- | ||
| - | | | + | | W3110 – Reinvigorated |
|- | |- | ||
| - | | | + | | W3110 – Known Competent |
|- | |- | ||
|} | |} | ||
| Line 90: | Line 91: | ||
'''Aim - Results from Transformation of pTop''' | '''Aim - Results from Transformation of pTop''' | ||
| - | '''Results:''' Table below indicates there was significant growth from the Original | + | '''Results:''' Table below indicates there was significant growth from the Original W3110 cells, but not from other cell lines. |
| Line 97: | Line 98: | ||
! colspan="2" |Plates || Colony Formation | ! colspan="2" |Plates || Colony Formation | ||
|- | |- | ||
| - | | rowspan="3" | Protocol 1 || | + | | rowspan="3" | Protocol 1 || W3110 – Original || Yes |
|- | |- | ||
| - | | | + | | W3110 – Reinvigorated || No |
|- | |- | ||
| - | | | + | | W3110 – Known Competent|| No |
|- | |- | ||
| - | | rowspan="3" | Protocol 2 || | + | | rowspan="3" | Protocol 2 || W3110 – Original || Yes |
|- | |- | ||
| - | | | + | | W3110 – Reinvigorated || No |
|- | |- | ||
| - | | | + | | W3110 – Known Competent || No |
|- | |- | ||
|} | |} | ||
| - | '''Conclusion:''' The Original Cell line appears to be competent for both Protocols, while our reinvigorated cell line does not appear to be competent with either. The cells known to be competent showed no growth, for reasons that are unclear. For our Original | + | '''Conclusion:''' The Original Cell line appears to be competent for both Protocols, while our reinvigorated cell line does not appear to be competent with either. The cells known to be competent showed no growth, for reasons that are unclear. For our Original W3110 cell line, there was greater colony formation with Protocol 1 than with Protocol 2, so we shall proceed with this protocol. It is likely that the failure of our transformations is due to the switch to protocol 2, and the use of too little BioBrick to transform our cells. The protocol recommends 1-2ul, but members around the lab generally use 5ul for transforming cells. We will take this approach and see if our transformations improve. |
<html> | <html> | ||
</div><div class="experiment"></div> | </div><div class="experiment"></div> | ||
| - | <img src="https://static.igem.org/mediawiki/2012/8/8e/Ucl2012-labbook-graph6-3.png" /><div class="experimentContent"> | + | <img id="6-3" src="https://static.igem.org/mediawiki/2012/8/8e/Ucl2012-labbook-graph6-3.png" /><div class="experimentContent"> |
</html> | </html> | ||
| Line 123: | Line 124: | ||
Method: | Method: | ||
| - | + | <html><div class="protocol protocol-Transformation">Transformation Protocol 1</div><div class="protocolContent"></html>{{:Team:University_College_London/Protocols/Transformation1}}<html></div></html> | |
| - | '''Step 1 – Thawing Cells:''' Use | + | '''Step 1 – Thawing Cells:''' Use W3110 cell line created in Week 2 (Expt 2.1) |
'''Step 3 – Addition of BioBrick:''' To each 2ml eppendorf, add 1ul of the following BioBricks. Note: we have changed the protocol for our positive control. Previously it contained no BioBrick, but it has been recommended to us that we transform our positive control such that there is one for each BioBrick – this will tell us if the BioBrick has in any way affected cell viability. This will be used from this point onwards. Include an extra tube as a negative control, with no BioBrick added | '''Step 3 – Addition of BioBrick:''' To each 2ml eppendorf, add 1ul of the following BioBricks. Note: we have changed the protocol for our positive control. Previously it contained no BioBrick, but it has been recommended to us that we transform our positive control such that there is one for each BioBrick – this will tell us if the BioBrick has in any way affected cell viability. This will be used from this point onwards. Include an extra tube as a negative control, with no BioBrick added | ||
| Line 204: | Line 205: | ||
|- | |- | ||
|} | |} | ||
| + | |||
| + | {{:Team:University_College_London/templates/pictureinsert|title=Picture 1|url=images/5/57/UCLiGEM2012Agar_6.3.1.png}} | ||
Latest revision as of 01:07, 27 September 2012



Contents |
Tuesday 17.7.12
Aim -Testing the competency of the Reinvigorated Cell Line (W3110): After our original attempt to generate competency of the W3110 cell line was inadequate, we undertook a reinvigoration of the W310 cell line (Expt 5.2). Here we aim to test the competency of the reinvigorated cell line using the same protocol as used in Expt 5.3, by evaluating their growth after transformation with a plasmid of a known high concentration (297ng/ul).
Method
Step 1 - Thawing Cells: Thaw competent cells on ice.
Step 2 - Adding cells: Add 50 µL of thawed competent cells into pre-chilled 2ml tube.
Step 3 - Addition of BioBrick: Add 1 - 2 µL of the resuspended DNA to the 2ml tube. Pipette up and down a few times, gently. Make sure to keep the competent cells on ice.
Step 4 - Incubation: Close tube and incubate the cells on ice for 30 minutes.
Step 5 - Heat Shock: Heat shock the cells by immersion in a pre-heated water bath at 42ºC for 60 seconds.
Step 6 - Incubation: Incubate the cells on ice for 5 minutes.
Step 7 - Add media: Add 200 μl of SOC media or LB broth
Step 8 - Incubation: Incubate the cells at 37ºC for 2 hours while the tubes are rotating or shaking.
Step 9 - Label plates: Label two petri dishes with LB agar and the appropriate antibiotic(s) with the part number, plasmid backbone, and antibiotic resistance. Plate 20 µl and 200 µl of the transformation onto the dishes, and spread. This helps ensure that you will be able to pick out a single colony.
Step 10 - Culture:Incubate the plate at 37ºC for 12-14 hours, making sure the agar side of the plate is up. If incubated for too long the antibiotics start to break down and un-transformed cells will begin to grow. This is especially true for ampicillin - because the resistance enzyme is excreted by the bacteria, and inactivates the antibiotic outside of the bacteria.
Step 11 - Colony Picking: You can pick a single colony, make a glycerol stock, grow up a cell culture and miniprep.
Step 1 – Thawing Cells: Use the reinvigorated W3110 cell line created in Week 5 (Expt 5.2)
Step 3 – Addition of BioBrick: To one 2ml eppendorf, add 1ul of pTop plasmid, and to another add nothing – this will be a control.
Step 7 – Adding Broth: SOC media was used, as it is preferred for this protocol.
Step 8 - Incubation: The table below indicates the Ampicillin concentration of the Agar gels.
| Samples | Volume Innoculated | Antibiotic in Gel (conc) | |
|---|---|---|---|
| Plasmid | pTOP | 36ul | Ampicillin (50ug/ml) |
| Control | Positive (No Plasmid) | 36ul | No Antibiotic |
| Negative (No Plasid) | 36ul | Ampicillin (50ug/ml) | |
Wednesday (18.7.12)
Aim - Results from Transformation
Result: Table below indicates there was no growth for our cells, but that the controls worked. Also included is an image of each plate.
| Samples | Growth/No Growth | |
|---|---|---|
| Plasmid | pTOP | No Growth |
| Control | Positive (No Plasmid) | Growth |
| Negative (No Plasid) | No Growth | |
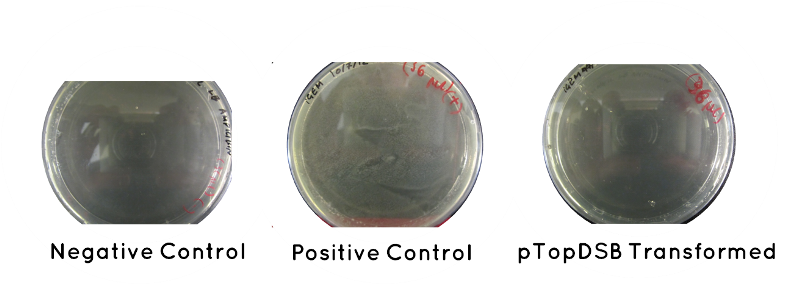

Wednesday (18.7.12)
Aim - Testing the transformation protocols: In light of the problems with cell competency, we decided to test whether the choice of protocol was contributing to the poor transformation results – especially as it would require another week to set up another cell line. We used a known competent cell line, and tested its competency against both of our cell lines, for Transformation Protocol 1 vs Transformation Protocol 2. Each sample was plated on an Ampicillin positive Agar and incubated overnight.
Step 1 - Addition of BioBrick: To the still frozen competent cells, add 1 - 5 µL of the resuspended DNA to the 2ml tube.
Step 4 - Incubation: Close tube and incubate the cells on ice for 45 minutes.
Step 5 - Heat Shock: Heat shock the cells by immersion in a pre-heated water bath at 37ºC for 10 minutes.
Step 6 - Incubation: Incubate the cells on ice for 2 minutes.
Step 7 - Add media: Add 1.5ml of Lysogeny Broth and transfer to a falcon tube.
Step 8 - Incubation: Incubate the cells at 37ºC for 1 hour at RPM 550.
Step 9 - Transfer: transfer the solution back into a 1.5ml Eppendorf and centrifuge
RPM: 14000
Time: 2 minutes
Temperature (18-25oC)
Step 10 - Resuspend:Remove supernatant and resuspend in 100ul LB
Step 11 - Plating: Spread the resuspended cell solution onto a selective nutrient agar plate. Place the plates in a 37°C static incubator, leave overnight (alternatively a 30°C static incubator over the weekend)
Step 1 - Thawing Cells: Thaw competent cells on ice.
Step 2 - Adding cells: Add 50 µL of thawed competent cells into pre-chilled 2ml tube.
Step 3 - Addition of BioBrick: Add 1 - 2 µL of the resuspended DNA to the 2ml tube. Pipette up and down a few times, gently. Make sure to keep the competent cells on ice.
Step 4 - Incubation: Close tube and incubate the cells on ice for 30 minutes.
Step 5 - Heat Shock: Heat shock the cells by immersion in a pre-heated water bath at 42ºC for 60 seconds.
Step 6 - Incubation: Incubate the cells on ice for 5 minutes.
Step 7 - Add media: Add 200 μl of SOC media or LB broth
Step 8 - Incubation: Incubate the cells at 37ºC for 2 hours while the tubes are rotating or shaking.
Step 9 - Label plates: Label two petri dishes with LB agar and the appropriate antibiotic(s) with the part number, plasmid backbone, and antibiotic resistance. Plate 20 µl and 200 µl of the transformation onto the dishes, and spread. This helps ensure that you will be able to pick out a single colony.
Step 10 - Culture:Incubate the plate at 37ºC for 12-14 hours, making sure the agar side of the plate is up. If incubated for too long the antibiotics start to break down and un-transformed cells will begin to grow. This is especially true for ampicillin - because the resistance enzyme is excreted by the bacteria, and inactivates the antibiotic outside of the bacteria.
Step 11 - Colony Picking: You can pick a single colony, make a glycerol stock, grow up a cell culture and miniprep.
Step ?: The table below describes the plates necessary for this investigation, each of which shoud carry Ampicillin antibiotic at a concentration of 50ug/ml.
| Plates | |
|---|---|
| Protocol 1 | W3110 – Original |
| W3110 – Reinvigorated | |
| W3110 – Known Competent | |
| Protocol 2 | W3110 – Original |
| W3110 – Reinvigorated | |
| W3110 – Known Competent | |
Thursday 19.7.12
Aim - Results from Transformation of pTop
Results: Table below indicates there was significant growth from the Original W3110 cells, but not from other cell lines.
| Plates | Colony Formation | |
|---|---|---|
| Protocol 1 | W3110 – Original | Yes |
| W3110 – Reinvigorated | No | |
| W3110 – Known Competent | No | |
| Protocol 2 | W3110 – Original | Yes |
| W3110 – Reinvigorated | No | |
| W3110 – Known Competent | No | |
Conclusion: The Original Cell line appears to be competent for both Protocols, while our reinvigorated cell line does not appear to be competent with either. The cells known to be competent showed no growth, for reasons that are unclear. For our Original W3110 cell line, there was greater colony formation with Protocol 1 than with Protocol 2, so we shall proceed with this protocol. It is likely that the failure of our transformations is due to the switch to protocol 2, and the use of too little BioBrick to transform our cells. The protocol recommends 1-2ul, but members around the lab generally use 5ul for transforming cells. We will take this approach and see if our transformations improve.

Thursday 19.7.12
Aim – Transformations: Expt 6.3 will be transforming our main Constitutive Promoter - BBa_J23119 – which is to be used in a number of modules. It will also aim to transform the Gas Vesicle Polycistronic Gene BBa_I750016, which is required for the production of gas vesicles. Finally, it will also transform the Ribosome Binding Site BBa_B0034, which failed to transform in Expt 5.1, and the Double Termination BBa_B0015. Both of these are key to all modules. Method:
Step 1 - Addition of BioBrick: To the still frozen competent cells, add 1 - 5 µL of the resuspended DNA to the 2ml tube.
Step 4 - Incubation: Close tube and incubate the cells on ice for 45 minutes.
Step 5 - Heat Shock: Heat shock the cells by immersion in a pre-heated water bath at 37ºC for 10 minutes.
Step 6 - Incubation: Incubate the cells on ice for 2 minutes.
Step 7 - Add media: Add 1.5ml of Lysogeny Broth and transfer to a falcon tube.
Step 8 - Incubation: Incubate the cells at 37ºC for 1 hour at RPM 550.
Step 9 - Transfer: transfer the solution back into a 1.5ml Eppendorf and centrifuge
RPM: 14000
Time: 2 minutes
Temperature (18-25oC)
Step 10 - Resuspend:Remove supernatant and resuspend in 100ul LB
Step 11 - Plating: Spread the resuspended cell solution onto a selective nutrient agar plate. Place the plates in a 37°C static incubator, leave overnight (alternatively a 30°C static incubator over the weekend)
Step 1 – Thawing Cells: Use W3110 cell line created in Week 2 (Expt 2.1)
Step 3 – Addition of BioBrick: To each 2ml eppendorf, add 1ul of the following BioBricks. Note: we have changed the protocol for our positive control. Previously it contained no BioBrick, but it has been recommended to us that we transform our positive control such that there is one for each BioBrick – this will tell us if the BioBrick has in any way affected cell viability. This will be used from this point onwards. Include an extra tube as a negative control, with no BioBrick added
| Function | Module | ||
|---|---|---|---|
| BioBrick | BBa_J23119 | Constitutive Promoter | Degradation/
Salt Tolerance/ Containment |
| BBa_I750016 | Gas Vesicle Polycistronic Gene | Buoyancy | |
| BBa_B0015 | Double Terminator | All | |
| BBa_B0034 | Ribosome Binding Site (RBS) | All | |
| Control | (No BioBricks) | ||
Step 9 – Plating samples on Agar Plates: The table below indicates the chosen inoculation volume (two for each BioBrick) and the correct gel antibiotic concentration for all samples.
| Samples | Volume Inoculated | Antibiotic in Gel (ug/ml) | |
|---|---|---|---|
| BioBrick | BBa_J23119 | 10ul | Ampicillin(50ug/ml) |
| 90ul | |||
| BBa_I750016 | 10ul | ||
| 90ul | |||
| BBa_B0015 | 10ul | ||
| 90ul | |||
| BBa_B0034 | 10ul | ||
| 90ul | |||
| Control | Positive Positive (Contains BioBrick – one for each of the above) | 36ul | No Antibiotic |
| Negative (No BioBrick) | 36ul | 1x Ampicillin(50ug/ml) | |
Friday 20.7.12
Aim 1 - Results of Transformation Result: Table below indicates there was growth for all of our transformed cells, and that the controls worked. Also included is an image of each plate.
| Samples | Volume Inoculated | Growth/ No Growth | |
|---|---|---|---|
| BioBrick | BBa_J23119 | 10ul | Growth |
| 90ul | Growth | ||
| BBa_I750016 | 10ul | Growth | |
| 90ul | Growth | ||
| BBa_B0015 | 10ul | Growth | |
| 90ul | Growth | ||
| BBa_B0034 | 10ul | Growth | |
| 90ul | Growth | ||
| Control | Positive Positive (Contains BioBrick – one for each of the above) | 36ul | Growth |
| Negative (No BioBrick) | 36ul | No Growth | |
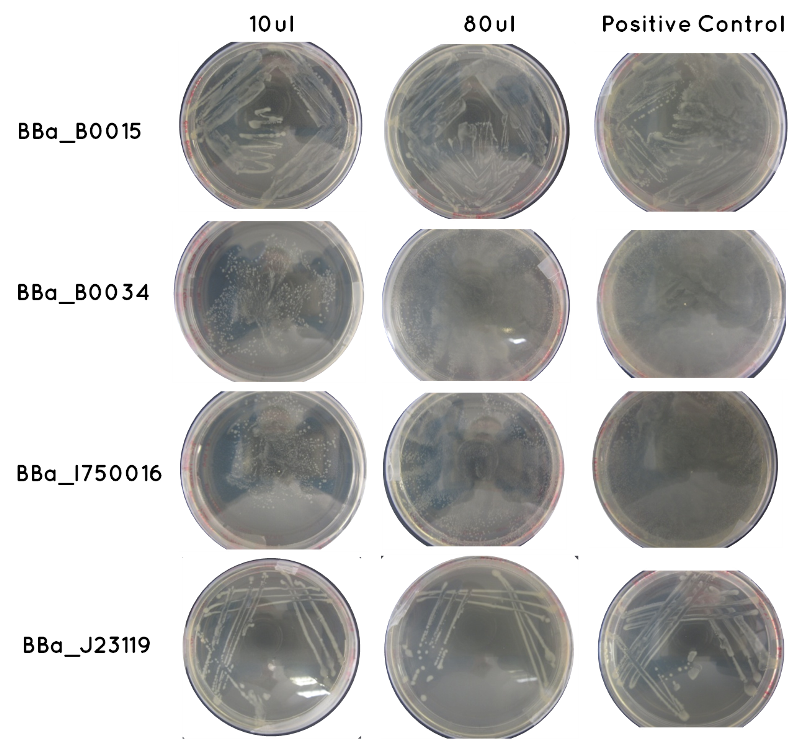
Conclusion: As of week 7 we can continue with colony picking for these agar plates, which will enable us to culture our transformed cells, and purify the plasmid. The purified plasmid will undergo restriction digest to determine the presence of the correct BioBrick inserts.
 "
"