Team:University College London/LabBook/Week17
From 2012.igem.org
YanikaBorg (Talk | contribs) (→17-3) |
(→17-3) |
||
| (24 intermediate revisions not shown) | |||
| Line 7: | Line 7: | ||
| - | We used sonication to disrupt three cell types: unmodified W3110 strain E. coli and W3110 strains harbouring chloramphenical resistance plasmids encoding a laccase (BBa_K729006) or the DsbA-Nuclease (BBa_K729019). All three strains were grown to OD600= 2.0 prior to sonication, in 2mL LB broth which contained 100ug/mL chloramphenical for plasmid-harbouring cells. | + | |
| + | '''Aim:''' To determine the ability of BBa_K729019 to act as a biosafety device. | ||
| + | |||
| + | |||
| + | '''Method:''' We used sonication to disrupt three cell types: unmodified W3110 strain E. coli and W3110 strains harbouring chloramphenical resistance plasmids encoding a laccase (BBa_K729006) or the DsbA-Nuclease (BBa_K729019). All three strains were grown to OD600= 2.0 prior to sonication, in 2mL LB broth which contained 100ug/mL chloramphenical for plasmid-harbouring cells. | ||
| Line 16: | Line 20: | ||
| - | As expected, transformation of TOP10 cells with disruptate of unmodified W3110 did not yield any cells able to grow on chloramphenicol plates (FigXA). In contrast transformation with disruptate of W3110 harbouring the BBa_K729006 laccase plasmid yielded many TOP10 colonies on chloramphenicol plates (FigXA), indicating that plasmid DNA in the disruptate was able to transform TOP10 cells. | + | '''Results:''' As expected, transformation of TOP10 cells with disruptate of unmodified W3110 did not yield any cells able to grow on chloramphenicol plates (FigXA). In contrast transformation with disruptate of W3110 harbouring the BBa_K729006 laccase plasmid yielded many TOP10 colonies on chloramphenicol plates (FigXA), indicating that plasmid DNA in the disruptate was able to transform TOP10 cells. |
| Line 22: | Line 26: | ||
| - | We suggest periplasmic nuclease expression provides a promising strategy for preventing transfer of genetically modified DNA. This is particularly valuable for the application of synthetic biology in environmental contexts. | + | |
| + | '''Conclusion:''' We suggest periplasmic nuclease expression provides a promising strategy for preventing transfer of genetically modified DNA. This is particularly valuable for the application of synthetic biology in environmental contexts. | ||
<html> | <html> | ||
</div><div class="experiment"></div> | </div><div class="experiment"></div> | ||
| Line 28: | Line 33: | ||
<div class="experimentContent"></html> | <div class="experimentContent"></html> | ||
| - | == | + | ==Growth comparison of nuclease positive and nuclease negative cells== |
| + | =Aim= | ||
| + | To determine if nuclease was toxic to our cells. | ||
| + | =Method= | ||
| + | The following procedure must be repeated for bothfor nuclease positive and nuclease negative BL21 cells. Nuclease negative cells act as a control. | ||
| + | 1. In 150ml of LB inoculate 10 ul of cells | ||
| + | 2. Split into three separate flasks (50ml in three 250ml flasks) | ||
| + | 3. Grow in 37 degrees, 200 rpm | ||
| + | 4. Take OD measurement every hour for 12 hours | ||
| + | 5. Repeat inoculation and measurements three times. | ||
| Line 72: | Line 86: | ||
|- | |- | ||
|} | |} | ||
| + | |||
| + | |||
| + | {| class="bigtable" | ||
| + | |- | ||
| + | ! colspan="7" style="background: #efefef;" | Second Inocculation | ||
| + | |- | ||
| + | ! x | ||
| + | ! colspan="3" | Nuclease positive | ||
| + | ! colspan="3" | Nuclease negative (control) | ||
| + | |- | ||
| + | |Time || 1|| 2|| 3 || 1|| 2|| 3 | ||
| + | |- | ||
| + | |0||0.001 ||0.001 ||0.001 ||0.001 ||0.001 ||0.002 | ||
| + | |- | ||
| + | |1||0.004 ||0.006 ||0.006 ||0.003 ||0.002 ||0.004 | ||
| + | |- | ||
| + | |2||0.006 ||0.009 ||0.008 ||0.008 ||0.006 ||0.01 | ||
| + | |- | ||
| + | |3||0.009 ||0.014 ||0.011 ||0.026 ||0.017 ||0.023 | ||
| + | |- | ||
| + | |4||0.028 ||0.029 ||0.028 ||0.034 ||0.029 ||0.034 | ||
| + | |- | ||
| + | |5||0.048 ||0.07 ||0.066 ||0.068 ||0.066 ||0.072 | ||
| + | |- | ||
| + | |6||0.079 ||0.111 ||0.107 ||0.092 ||0.096 ||0.099 | ||
| + | |- | ||
| + | |7||0.181 ||0.207 ||0.203 ||0.179 ||0.185 ||0.191 | ||
| + | |- | ||
| + | |8||0.28 ||0.47 ||0.36 ||0.311 ||0.289 ||0.298 | ||
| + | |- | ||
| + | |9||0.689 ||0.919 ||0.8 ||0.671 ||0.672 ||0.683 | ||
| + | |- | ||
| + | |10||0.798 ||1.491 ||1.299 ||0.781 ||0.788 ||0.795 | ||
| + | |- | ||
| + | |11||0.909 ||1.8 ||1.664 ||0.924 ||0.974 ||0.981 | ||
| + | |- | ||
| + | |12||1.018 ||2.038 ||1.892 ||1.073 ||1.083 ||1.097 | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | |||
| + | {| class="bigtable" | ||
| + | |- | ||
| + | ! colspan="7" style="background: #efefef;" | Third Inocculation | ||
| + | |- | ||
| + | ! x | ||
| + | ! colspan="3" | Nuclease positive | ||
| + | ! colspan="3" | Nuclease negative (control) | ||
| + | |- | ||
| + | |Time || 1|| 2|| 3 || 1|| 2|| 3 | ||
| + | |- | ||
| + | |0||0.002 ||0.001 ||0.002 ||0.001 ||0.001 ||0.001 | ||
| + | |- | ||
| + | |1||0.005 ||0.003 ||0.005 ||0.002 ||0.004 ||0.003 | ||
| + | |- | ||
| + | |2||0.007 ||0.006 ||0.007 ||0.007 ||0.005 ||0.006 | ||
| + | |- | ||
| + | |3||0.011 ||0.016 ||0.01 ||0.018 ||0.014 ||0.017 | ||
| + | |- | ||
| + | |4||0.029 ||0.033 ||0.027 ||0.027 ||0.023 ||0.025 | ||
| + | |- | ||
| + | |5||0.05 ||0.08 ||0.049 ||0.063 ||0.051 ||0.057 | ||
| + | |- | ||
| + | |6||0.087 ||0.116 ||0.085 ||0.091 ||0.083 ||0.081 | ||
| + | |- | ||
| + | |7||0.185 ||0.211 ||0.187 ||0.17 ||0.184 ||0.178 | ||
| + | |- | ||
| + | |8||0.309 ||0.481 ||0.306 ||0.291 ||0.297 ||0.29 | ||
| + | |- | ||
| + | |9||0.681 ||0.924 ||0.799 ||0.666 ||0.686 ||0.68 | ||
| + | |- | ||
| + | |10||0.806 ||1.502 ||0.987 ||0.773 ||0.79 ||0.782 | ||
| + | |- | ||
| + | |11||0.917 ||1.811 ||0.969 ||0.914 ||0.95 ||0.941 | ||
| + | |- | ||
| + | |12||1.031 ||2.049 ||1.159 ||1.064 ||1.099 ||1.093 | ||
| + | |- | ||
| + | |} | ||
| + | |||
<html></div> | <html></div> | ||
| Line 79: | Line 172: | ||
== 17-4== | == 17-4== | ||
| + | |||
| + | '''Aim''' To investigate if presence of irrE in W3110 improroves growth in MB (Marine Broth)in comparison to wild type W3110 growth in MB. | ||
| + | |||
| + | '''Method''' | ||
| + | |||
| + | Five different samples: 1)LB + W3110; 2)LB + Indolifex; 3)MB + W3110 4)MB + Indolifex; 5)MB+W3110 (irrE present); | ||
| + | The following procedure refers to each of the samples above: | ||
| + | |||
| + | |||
| + | 1. 10ul of cells (W3110/Indolifex/W3110+irrE) from glycerol stockinto 150ml of MB/LB | ||
| + | |||
| + | 2. Split this into three flasks (in each 250ml flask add 50ml of inoculate) | ||
| + | |||
| + | 3. Take OD measurements every hour for 9 hours. | ||
| + | |||
| + | |||
| + | |||
| + | ''' LB + W3110 ''' | ||
| Line 105: | Line 216: | ||
| 9 || 0.669|| 0.678 || 0.675 | | 9 || 0.669|| 0.678 || 0.675 | ||
|} | |} | ||
| + | |||
| + | |||
| + | ''' LB + Indolifex ''' | ||
| + | |||
| + | {| class="bigtable" | ||
| + | |- | ||
| + | ! Time !! Run 1 !! Run 2 !! Run3 | ||
| + | |- | ||
| + | | 0 || 0.001 || 0.001 || 0.001 | ||
| + | |- | ||
| + | | 1 || 0.002 || 0.002 ||0.001 | ||
| + | |- | ||
| + | | 2 || 0.004|| 0.003|| 0.004 | ||
| + | |- | ||
| + | | 3 || 0.009|| 0.009|| 0.008 | ||
| + | |- | ||
| + | | 4 || 0.019|| 0.017|| 0.017 | ||
| + | |- | ||
| + | | 5 || 0.038|| 0.036|| 0.037 | ||
| + | |- | ||
| + | | 6 || 0.059|| 0.064 || 0.055 | ||
| + | |- | ||
| + | | 7 || 0.077|| 0.082 || 0.072 | ||
| + | |- | ||
| + | | 8 || 0.138|| 0.136 || 0.129 | ||
| + | |- | ||
| + | | 9 || 0.222|| 0.221 || 0.217 | ||
| + | |} | ||
| + | |||
| + | |||
| + | '''MB + W3110''' | ||
| + | |||
| + | {| class="bigtable" | ||
| + | |- | ||
| + | ! Time !! Run 1 !! Run 2 !! Run3 | ||
| + | |- | ||
| + | | 0 || 0.001 || 0.001 || 0.001 | ||
| + | |- | ||
| + | | 1 || 0.001 || 0.001 || 0.001 | ||
| + | |- | ||
| + | | 2 || 0.001 || 0.001 || 0.001 | ||
| + | |- | ||
| + | | 3 || 0.005|| 0.004|| 0.005 | ||
| + | |- | ||
| + | | 4 || 0.007|| 0.006|| 0.006 | ||
| + | |- | ||
| + | | 5 || 0.015|| 0.012|| 0.014 | ||
| + | |- | ||
| + | | 6 || 0.032|| 0.029|| 0.03 | ||
| + | |- | ||
| + | | 7 || 0.049|| 0.043 || 0.045 | ||
| + | |- | ||
| + | | 8 || 0.061|| 0.057|| 0.059 | ||
| + | |- | ||
| + | | 9 || 0.072|| 0.068 || 0.06 | ||
| + | |} | ||
| + | |||
| + | |||
| + | '''MB + Indolifex''' | ||
| + | |||
| + | {| class="bigtable" | ||
| + | |- | ||
| + | ! Time !! Run 1 !! Run 2 !! Run3 | ||
| + | |- | ||
| + | | 0 || 0.001 || 0.001 || 0.001 | ||
| + | |- | ||
| + | | 1 || 0.004 || 0.004 || 0.004 | ||
| + | |- | ||
| + | | 2 || 0.012 || 0.011 || 0.012 | ||
| + | |- | ||
| + | | 3 || 0.029|| 0.027|| 0.028 | ||
| + | |- | ||
| + | | 4 || 0.038|| 0.036|| 0.36 | ||
| + | |- | ||
| + | | 5 || 0.072|| 0.067|| 0.07 | ||
| + | |- | ||
| + | | 6 || 0.091|| 0.088|| 0.09 | ||
| + | |- | ||
| + | | 7 || 0.179|| 0.17 || 0.174 | ||
| + | |- | ||
| + | | 8 || 0.339|| 0.3|| 0.344 | ||
| + | |- | ||
| + | | 9 || 0.774|| 0.75|| 0.766 | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | ''' MB+W3110 (irrE present)''' | ||
| + | |||
| + | {| class="bigtable" | ||
| + | |- | ||
| + | ! Time !! Run 1 !! Run 2 !! Run3 | ||
| + | |- | ||
| + | | 0 || 0.001 || 0.001 || 0.001 | ||
| + | |- | ||
| + | | 1 || 0.002 || 0.002 || 0.003 | ||
| + | |- | ||
| + | | 2 || 0.008 || 0.007 || 0.009 | ||
| + | |- | ||
| + | | 3 || 0.021|| 0.019|| 0.02 | ||
| + | |- | ||
| + | | 4 || 0.032|| 0.03|| 0.3 | ||
| + | |- | ||
| + | | 5 || 0.061|| 0.06|| 0.063 | ||
| + | |- | ||
| + | | 6 || 0.085|| 0.086|| 0.089 | ||
| + | |- | ||
| + | | 7 || 0.164|| 0.166 || 0.165 | ||
| + | |- | ||
| + | | 8 || 0.29|| 0.293|| 0.289 | ||
| + | |- | ||
| + | | 9 || 0.639|| 0.644|| 0.65 | ||
| + | |} | ||
| + | |||
| + | ''' Conclusion''' from the tables above we can see that irrE positive W3110 cells have greater growth in comparison to wild type W3110 cells in MB, therefore we can conclude that the addition of irrE improves W3110 growth in marine medium. | ||
| + | |||
<html> | <html> | ||
</div><div class="experiment"></div></html> | </div><div class="experiment"></div></html> | ||
{{:Team:University_College_London/templates/foot}} | {{:Team:University_College_London/templates/foot}} | ||
Latest revision as of 01:51, 27 October 2012



Contents |
Material from cells containing our Biosafety BioBrick is not able to transform commercial competent cells
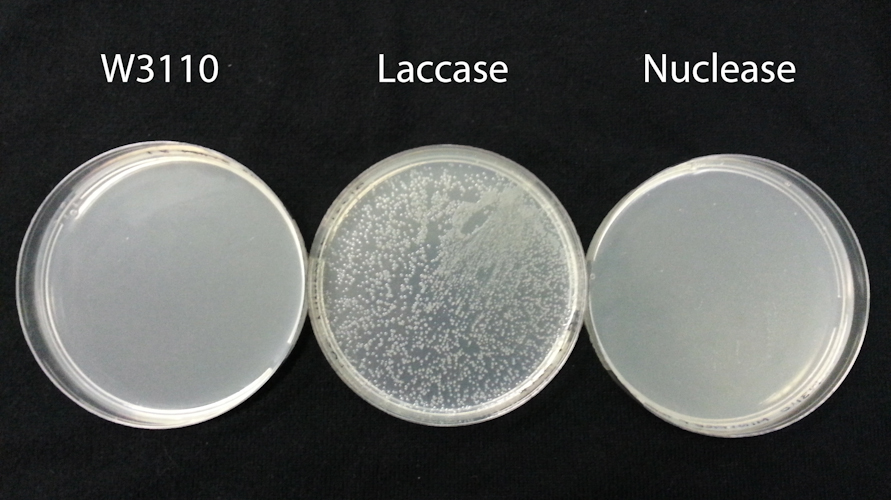
Aim: To determine the ability of BBa_K729019 to act as a biosafety device.
Method: We used sonication to disrupt three cell types: unmodified W3110 strain E. coli and W3110 strains harbouring chloramphenical resistance plasmids encoding a laccase (BBa_K729006) or the DsbA-Nuclease (BBa_K729019). All three strains were grown to OD600= 2.0 prior to sonication, in 2mL LB broth which contained 100ug/mL chloramphenical for plasmid-harbouring cells.
After sonication, disruptates were incubated for 10min at room temperature. 5uL of the disruptate was used to transform an aliquot of TOP10 chemically competent cells, following manufacturer instructions. As a control, we also spread 20uL of the disruptate material directly onto LB agar plates with and without 100ug/mL Chloramphenical. No colonies were observed on the control plates for any of the three strains (data not shown), indicating sonication had successfully disrupted all cells.
Results: As expected, transformation of TOP10 cells with disruptate of unmodified W3110 did not yield any cells able to grow on chloramphenicol plates (FigXA). In contrast transformation with disruptate of W3110 harbouring the BBa_K729006 laccase plasmid yielded many TOP10 colonies on chloramphenicol plates (FigXA), indicating that plasmid DNA in the disruptate was able to transform TOP10 cells.
Transformation with disruptate of W3110 harbouring the BBa_K729019 nuclease plasmid yielded no colonies on chloramphenicol plates (FigXC). BBa_K729004 differs from BBa_K729006 only in that it contains an ORF encoding the periplasmic nuclease instead of laccase. As such, we conclude the periplasmic nuclease has sufficient DNAase activity in the disruptate to reduce the amount of plasmid DNA present to below the threshold capable of transforming TOP10 cells.
Conclusion: We suggest periplasmic nuclease expression provides a promising strategy for preventing transfer of genetically modified DNA. This is particularly valuable for the application of synthetic biology in environmental contexts.

Growth comparison of nuclease positive and nuclease negative cells
Aim
To determine if nuclease was toxic to our cells.
Method
The following procedure must be repeated for bothfor nuclease positive and nuclease negative BL21 cells. Nuclease negative cells act as a control.
1. In 150ml of LB inoculate 10 ul of cells 2. Split into three separate flasks (50ml in three 250ml flasks) 3. Grow in 37 degrees, 200 rpm 4. Take OD measurement every hour for 12 hours 5. Repeat inoculation and measurements three times.
| First Inocculation | ||||||
|---|---|---|---|---|---|---|
| x | Nuclease positive | Nuclease negative (control) | ||||
| Time | 1 | 2 | 3 | 1 | 2 | 3 |
| 0 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 |
| 1 | 0.003 | 0.005 | 0.004 | 0.003 | 0.003 | 0.004 |
| 2 | 0.005 | 0.008 | 0.005 | 0.009 | 0.005 | 0.008 |
| 3 | 0.008 | 0.011 | 0.009 | 0.02 | 0.014 | 0.018 |
| 4 | 0.021 | 0.03 | 0.024 | 0.03 | 0.026 | 0.028 |
| 5 | 0.053 | 0.076 | 0.059 | 0.064 | 0.057 | 0.066 |
| 6 | 0.084 | 0.116 | 0.086 | 0.08 | 0.088 | 0.89 |
| 7 | 0.183 | 0.217 | 0.189 | 0.18 | 0.188 | 0.179 |
| 8 | 0.299 | 0.479 | 0.303 | 0.306 | 0.297 | 0.292 |
| 9 | 0.684 | 0.918 | 0.69 | 0.667 | 0.678 | 0.669 |
| 10 | 0.801 | 1.499 | 0.807 | 0.787 | 0.799 | 0.777 |
| 11 | 0.913 | 1.804 | 0.92 | 0.921 | 0.955 | 0.969 |
| 12 | 1.022 | 2.033 | 1.034 | 1.079 | 1.111 | 1.121 |
| Second Inocculation | ||||||
|---|---|---|---|---|---|---|
| x | Nuclease positive | Nuclease negative (control) | ||||
| Time | 1 | 2 | 3 | 1 | 2 | 3 |
| 0 | 0.001 | 0.001 | 0.001 | 0.001 | 0.001 | 0.002 |
| 1 | 0.004 | 0.006 | 0.006 | 0.003 | 0.002 | 0.004 |
| 2 | 0.006 | 0.009 | 0.008 | 0.008 | 0.006 | 0.01 |
| 3 | 0.009 | 0.014 | 0.011 | 0.026 | 0.017 | 0.023 |
| 4 | 0.028 | 0.029 | 0.028 | 0.034 | 0.029 | 0.034 |
| 5 | 0.048 | 0.07 | 0.066 | 0.068 | 0.066 | 0.072 |
| 6 | 0.079 | 0.111 | 0.107 | 0.092 | 0.096 | 0.099 |
| 7 | 0.181 | 0.207 | 0.203 | 0.179 | 0.185 | 0.191 |
| 8 | 0.28 | 0.47 | 0.36 | 0.311 | 0.289 | 0.298 |
| 9 | 0.689 | 0.919 | 0.8 | 0.671 | 0.672 | 0.683 |
| 10 | 0.798 | 1.491 | 1.299 | 0.781 | 0.788 | 0.795 |
| 11 | 0.909 | 1.8 | 1.664 | 0.924 | 0.974 | 0.981 |
| 12 | 1.018 | 2.038 | 1.892 | 1.073 | 1.083 | 1.097 |
| Third Inocculation | ||||||
|---|---|---|---|---|---|---|
| x | Nuclease positive | Nuclease negative (control) | ||||
| Time | 1 | 2 | 3 | 1 | 2 | 3 |
| 0 | 0.002 | 0.001 | 0.002 | 0.001 | 0.001 | 0.001 |
| 1 | 0.005 | 0.003 | 0.005 | 0.002 | 0.004 | 0.003 |
| 2 | 0.007 | 0.006 | 0.007 | 0.007 | 0.005 | 0.006 |
| 3 | 0.011 | 0.016 | 0.01 | 0.018 | 0.014 | 0.017 |
| 4 | 0.029 | 0.033 | 0.027 | 0.027 | 0.023 | 0.025 |
| 5 | 0.05 | 0.08 | 0.049 | 0.063 | 0.051 | 0.057 |
| 6 | 0.087 | 0.116 | 0.085 | 0.091 | 0.083 | 0.081 |
| 7 | 0.185 | 0.211 | 0.187 | 0.17 | 0.184 | 0.178 |
| 8 | 0.309 | 0.481 | 0.306 | 0.291 | 0.297 | 0.29 |
| 9 | 0.681 | 0.924 | 0.799 | 0.666 | 0.686 | 0.68 |
| 10 | 0.806 | 1.502 | 0.987 | 0.773 | 0.79 | 0.782 |
| 11 | 0.917 | 1.811 | 0.969 | 0.914 | 0.95 | 0.941 |
| 12 | 1.031 | 2.049 | 1.159 | 1.064 | 1.099 | 1.093 |

17-4
Aim To investigate if presence of irrE in W3110 improroves growth in MB (Marine Broth)in comparison to wild type W3110 growth in MB.
Method
Five different samples: 1)LB + W3110; 2)LB + Indolifex; 3)MB + W3110 4)MB + Indolifex; 5)MB+W3110 (irrE present); The following procedure refers to each of the samples above:
1. 10ul of cells (W3110/Indolifex/W3110+irrE) from glycerol stockinto 150ml of MB/LB
2. Split this into three flasks (in each 250ml flask add 50ml of inoculate)
3. Take OD measurements every hour for 9 hours.
LB + W3110
| Time | Run 1 | Run 2 | Run 3 |
|---|---|---|---|
| 0 | 0.001 | 0.001 | 0.001 |
| 1 | 0.002 | 0.003 | 0.004 |
| 2 | 0.008 | 0.007 | 0.008 |
| 3 | 0.02 | 0.019 | 0.019 |
| 4 | 0.029 | 0.026 | 0.027 |
| 5 | 0.06 | 0.059 | 0.062 |
| 6 | 0.087 | 0.089 | 0.089 |
| 7 | 0.184 | 0.185 | 0.183 |
| 8 | 0.302 | 0.297 | 0.292 |
| 9 | 0.669 | 0.678 | 0.675 |
LB + Indolifex
| Time | Run 1 | Run 2 | Run3 |
|---|---|---|---|
| 0 | 0.001 | 0.001 | 0.001 |
| 1 | 0.002 | 0.002 | 0.001 |
| 2 | 0.004 | 0.003 | 0.004 |
| 3 | 0.009 | 0.009 | 0.008 |
| 4 | 0.019 | 0.017 | 0.017 |
| 5 | 0.038 | 0.036 | 0.037 |
| 6 | 0.059 | 0.064 | 0.055 |
| 7 | 0.077 | 0.082 | 0.072 |
| 8 | 0.138 | 0.136 | 0.129 |
| 9 | 0.222 | 0.221 | 0.217 |
MB + W3110
| Time | Run 1 | Run 2 | Run3 |
|---|---|---|---|
| 0 | 0.001 | 0.001 | 0.001 |
| 1 | 0.001 | 0.001 | 0.001 |
| 2 | 0.001 | 0.001 | 0.001 |
| 3 | 0.005 | 0.004 | 0.005 |
| 4 | 0.007 | 0.006 | 0.006 |
| 5 | 0.015 | 0.012 | 0.014 |
| 6 | 0.032 | 0.029 | 0.03 |
| 7 | 0.049 | 0.043 | 0.045 |
| 8 | 0.061 | 0.057 | 0.059 |
| 9 | 0.072 | 0.068 | 0.06 |
MB + Indolifex
| Time | Run 1 | Run 2 | Run3 |
|---|---|---|---|
| 0 | 0.001 | 0.001 | 0.001 |
| 1 | 0.004 | 0.004 | 0.004 |
| 2 | 0.012 | 0.011 | 0.012 |
| 3 | 0.029 | 0.027 | 0.028 |
| 4 | 0.038 | 0.036 | 0.36 |
| 5 | 0.072 | 0.067 | 0.07 |
| 6 | 0.091 | 0.088 | 0.09 |
| 7 | 0.179 | 0.17 | 0.174 |
| 8 | 0.339 | 0.3 | 0.344 |
| 9 | 0.774 | 0.75 | 0.766 |
MB+W3110 (irrE present)
| Time | Run 1 | Run 2 | Run3 |
|---|---|---|---|
| 0 | 0.001 | 0.001 | 0.001 |
| 1 | 0.002 | 0.002 | 0.003 |
| 2 | 0.008 | 0.007 | 0.009 |
| 3 | 0.021 | 0.019 | 0.02 |
| 4 | 0.032 | 0.03 | 0.3 |
| 5 | 0.061 | 0.06 | 0.063 |
| 6 | 0.085 | 0.086 | 0.089 |
| 7 | 0.164 | 0.166 | 0.165 |
| 8 | 0.29 | 0.293 | 0.289 |
| 9 | 0.639 | 0.644 | 0.65 |
Conclusion from the tables above we can see that irrE positive W3110 cells have greater growth in comparison to wild type W3110 cells in MB, therefore we can conclude that the addition of irrE improves W3110 growth in marine medium.
 "
"