Team:University College London/LabBook/Week5
From 2012.igem.org
Sednanalien (Talk | contribs) (→Wednesday 11.7.12) |
Sednanalien (Talk | contribs) (→Monday 16.7.12) |
||
| (7 intermediate revisions not shown) | |||
| Line 151: | Line 151: | ||
|- | |- | ||
|} | |} | ||
| + | |||
== Tuesday (10.7.12) == | == Tuesday (10.7.12) == | ||
| Line 219: | Line 220: | ||
| 200ul | | 200ul | ||
|} | |} | ||
| + | |||
== Wednesday (11.7.12) == | == Wednesday (11.7.12) == | ||
| Line 277: | Line 279: | ||
| 200ul | | 200ul | ||
|} | |} | ||
| + | |||
== Thursday 12.7.12 == | == Thursday 12.7.12 == | ||
| Line 302: | Line 305: | ||
| 2x 200ul | | 2x 200ul | ||
|} | |} | ||
| + | |||
== Friday (13.7.12) == | == Friday (13.7.12) == | ||
| Line 319: | Line 323: | ||
'''Aim – Overnight Culture of Original line of W3110 cells:''' For the Reinvigoration we will be reapplying the protocol for Generating Competent Cells on the failed cell line. To do so, we must first inoculate the original cells into a falcon, and culture overnight. | '''Aim – Overnight Culture of Original line of W3110 cells:''' For the Reinvigoration we will be reapplying the protocol for Generating Competent Cells on the failed cell line. To do so, we must first inoculate the original cells into a falcon, and culture overnight. | ||
| + | |||
== Tuesday (10.7.12) == | == Tuesday (10.7.12) == | ||
| Line 327: | Line 332: | ||
<html><div class="protocol protocol-Competency">Day 1 of Generating Competent Cells</div><div class="protocolContent"></html>{{:Team:University_College_London/Protocols/Competency}}<html></div></html> | <html><div class="protocol protocol-Competency">Day 1 of Generating Competent Cells</div><div class="protocolContent"></html>{{:Team:University_College_London/Protocols/Competency}}<html></div></html> | ||
| + | |||
== Wednesday (11.7.12) == | == Wednesday (11.7.12) == | ||
| Line 334: | Line 340: | ||
<html><div class="protocol protocol-Competency">Day 2 of Generating Competent Cells</div><div class="protocolContent"></html>{{:Team:University_College_London/Protocols/Competency2}}<html></div></html> | <html><div class="protocol protocol-Competency">Day 2 of Generating Competent Cells</div><div class="protocolContent"></html>{{:Team:University_College_London/Protocols/Competency2}}<html></div></html> | ||
| + | |||
== Thursday (11.7.12) == | == Thursday (11.7.12) == | ||
| Line 374: | Line 381: | ||
| Negative (No plasmid) || 36ul || Ampicillin (50ug/ml) | | Negative (No plasmid) || 36ul || Ampicillin (50ug/ml) | ||
|} | |} | ||
| + | |||
== Monday 16.7.12 == | == Monday 16.7.12 == | ||
Latest revision as of 01:07, 27 September 2012



Contents |
Tuesday 10.7.12
Aim - Analytical Restriction Digest: This is intended to diagnose whether the correct BioBrick (BBa_I13522 and BBa_K540000) had been transformed into our bacteria, by measuring the size of the digested products against a DNA ladder.
Methods
Step 1 - Thawing cells: Thaw all materials on ice
Step 2 - Adding Ingredient: Add the following ingredients to autoclaved/sterile eppendorf tubes
| Component | Amount (ul) (one enzyme used) | Amount (ul) (two enzymes used) |
|---|---|---|
| dH20 | 2.5 | 1.5 |
| Buffer 1x | 1 | 1 |
| DNA template | 5 | 5 |
| BSA | 0.5 | 0.5 |
| Enzyme 1 | 1 | 2 |
| Enzyme 2 | N/A | 1 |
Step 3 - Addition of BioBrick: Flick contents gently and centrifuge.
Step 4 - Centrifuge:
RPM: 14000
Time: 1 minute
Temperature: 18oC
Step 5 - Digest Program: Place the samples on a thermocycler under the following conditions:
RPM: 550
Time: 2.5 hours
Temperature: 37oC
Step 6 - Denaturing Enzymes: If you are not running the samples on a gel immediately, denature the restriction enzymes by running the samples on a thermocycler under the following conditions:
RPM: 550
Time: 25 minutes
Temperature: 65oC
Step 2 – Setting up Digests and Controls: The protocol describes the recipe for (i) Digested Plasmid and (ii) Uncut Control. The table below indicates that an uncut and an EcoR1/Pst1 digested sample be set up for each BioBrick.
| Samples | Recipe | Enzyme Used! | |
|---|---|---|---|
| BioBrick | BBa_I13522 | Digested Plasmid | EcoR1 and Pst1 |
| Undigested Plasmid | None | ||
| BBa_K540000 | Digested Plasmid | EcoR1 and Pst1 | |
| Undigested Plasmid | None | ||
Preparing the Gel
Step 1: Within a conical flask, add 3ml 50X TAE, 1.5g Agarose, and 150ml RO water
Step 2: Heat for 1 min in microwave. Swirl. Heat again for 30s. If solution is clear stop. Else repeat.
Step 3: Cool solution under running cold water.
Step 4: Add 20ul ethidium bromide (normal concentration of EB solution is 500ug/ul)
Step 5: Pour into a sealed casting tray in a slow steady stream, ensuring there are no bubbles
Running a gel
Step 6: Add 1 part loading buffer to five parts of loading sample
Step 7: Position the gel in the tank and add TAE buffer, enough to cover the gel by several mm
Step 8: Add 5ul of DNA ladder to lane 1
Step 9: Add samples to the remaining wells
Step 10: Run at 100 volts for 1hour and 15 minutes
Imaging the Gel
Step 11: Place gel in GelDoc 2000 chamber
Step 12: Turn GelDoc 2000 chamber on
Step 13: From computer: Quantity One > Scanner > Gel_Doc_Xr>Manuqal Acquire
Step 14: Alter the exposure/settings to give a clear image.
TAE - Tris-acetate-EDTA
EDTA - ethylenediamine tetraacetic acid
Results: The image below shows a 1% Agarose Gel of an Analytical Restriction Enzyme Digest for Expt 4.1. The products shown by the gel are all incorrect. In lane 1 we expected a product equivalent to the combined size of the BBa_K540000 insert and pSB1C3 backbone (uncut plasmid). The position of this is shown by A, which corresponds to 5193 bp. In Lane 2 and Lane 3, there are no products, though we had expected to see a band corresponding to the size of the BBa_K540000 insert (3123bp) as shown by B and a band corresponding to the size of the pSB1C3 backbone (2070bp) as shown by C. In Lane 4, there should be a product corresponding to the combined size of the BBa_I13522 insert and pSB1A3 backbone (3092bp) which is indicated by D. This is absent. In lane 5, there are numerous products, but none corresponding to the size of the BBa_I13522 insert (937bp) as indicated by E, or the pSB1A3 backbone (2155bp) as indicated by F.
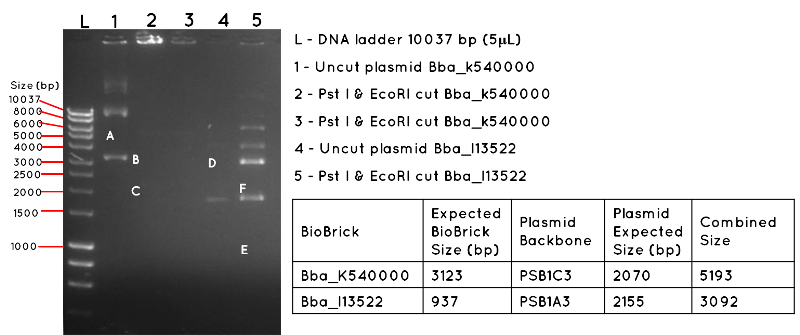
Conclusion: It is likely the products are the result of contamination, and the transformation was a failure
Software ND-1000 Model:
Step 1: Initialise the spectrophotometer by pipetting 1 µ of clean water onto lower optic surface, lowering the lever arm and selecting ‘initialise’ in the ND-1000 software
Step 2: Wipe and add elution buffer as negative control. Click blank in ND-1000 software
Step 3: Wipe and add 1 µl sample
Step 4: On the software set lambda to 260nm
Step 5: Lower the lever arm and click measure in ND-1000 software
Step 6: Take readings for concentration and purity
Step 7: Once measurement complete, wipe surface
Results: The table below indicates the concentrations indicated by the nanodropping for each plasmid.
| Header text | Concentration of plasmid (ng/ul) at wavelength 260 and 280 | |
|---|---|---|
| BioBrick | λ260 | λ280 |
| BBa_K540000 | 246.4 | 270.3 |
Wednesday 11.7.12
Aim - Repeat Analytical Digest for Expt 4.1 using different enzymes: This is intended to test again whether the correct plasmid had been transformed into our bacteria, after the last gel produced unusual results. This time we will use a different pairs of enzymes, which
Results: The image below shows a 1% Agarose Gel of an Analytical Restriction Enzyme Digest for Expt 4.1. Lane 1 displays the products of a single cut reaction, which for the plasmid carrying BBa_K540000 should produce a product of 5193 bp – the sum of the plasmid backbone pSB1C3 and the BBa_K540000 insert. A very faint band corresponding to this size is indicated by A, but the presence of more prominant bands of incorrect size suggests this reaction failed. Lane 2 displays a product of the size corresponding to the BBa_K540000 insert (3123bp) as shown by B, and a product corresponding the pSB1C3 backbone (2070 bp) as shown by C. The larger product, labelled D, could be excess uncut plasmid, as it corresponds approximately to the correct size (5193bp). Lane 3 shows the products of a single cut reaction for BBa_I13522. A product corresponding to the combined size of BBa_I13522 insert and pSB1A3 plasmid backbone is present (3092bp) as indicated by E, but presence of other bands is concerning. Lane 4 shows an absence of the correct size insert (937 bp) as indicated by G, and an absence of plasmid backbone pSB1A3 (2155bp), as indicated by F.

Conclusion: This gel would indicate that the BBa_K540000 transformation was a success, but there is some concern about the presence of extra bands in Lanes 1 and 2. For this reason we will repeat this gel when possible (See Expt 7.2). The BBa_I13522 transformation appears to be unsuccessful – there is not band corresponding to the insert. The digest will be repeated to be certain (See Expt 7.2).

Monday (9.7.12)
Aim: Transformation of Key BioBricks
Method
Step 1 - Thawing Cells: Thaw competent cells on ice.
Step 2 - Adding cells: Add 50 µL of thawed competent cells into pre-chilled 2ml tube.
Step 3 - Addition of BioBrick: Add 1 - 2 µL of the resuspended DNA to the 2ml tube. Pipette up and down a few times, gently. Make sure to keep the competent cells on ice.
Step 4 - Incubation: Close tube and incubate the cells on ice for 30 minutes.
Step 5 - Heat Shock: Heat shock the cells by immersion in a pre-heated water bath at 42ºC for 60 seconds.
Step 6 - Incubation: Incubate the cells on ice for 5 minutes.
Step 7 - Add media: Add 200 μl of SOC media or LB broth
Step 8 - Incubation: Incubate the cells at 37ºC for 2 hours while the tubes are rotating or shaking.
Step 9 - Label plates: Label two petri dishes with LB agar and the appropriate antibiotic(s) with the part number, plasmid backbone, and antibiotic resistance. Plate 20 µl and 200 µl of the transformation onto the dishes, and spread. This helps ensure that you will be able to pick out a single colony.
Step 10 - Culture:Incubate the plate at 37ºC for 12-14 hours, making sure the agar side of the plate is up. If incubated for too long the antibiotics start to break down and un-transformed cells will begin to grow. This is especially true for ampicillin - because the resistance enzyme is excreted by the bacteria, and inactivates the antibiotic outside of the bacteria.
Step 11 - Colony Picking: You can pick a single colony, make a glycerol stock, grow up a cell culture and miniprep.
Step 1 – Thawing Cells: Use W3110 cell line created in Week 2 (Expt 2.1)
Step 3 – Addition of BioBrick: To each 2ml eppendorf, add 1ul of the following BioBricks. Include an extra tube as a control, with no BioBrick added.
| Function | Module | ||
|---|---|---|---|
| BioBrick | BBa_K123003 | Oestrogen Receptor | Detection |
| BBa_K123002 | Oestrogen Response Element | Detection | |
| BBa_K398108 | Salt Tolerance Cluster | Salt Tolerance | |
| BBa_J23100 | Constitutive Promoter | ||
| BBa_J23107 | Constitutive Promoter | ||
| BBa_R0040 | TetR Repressible Promoter | Buoyancy | |
| BBa_J23106 | Constitutive Promoter | ||
| BBa_B0034 | Ribosome Binding Site (RBS) | All | |
| Control | (No BioBricks) | ||
Step 9 – Plating samples on Agar Plates: The table below indicates the chosen inoculation volume (two for each BioBrick) and the correct gel antibiotic concentration for all samples.
| Samples | Volume Inoculated | Antibiotic in Gel (ug/ml) | |
|---|---|---|---|
| BioBrick | BBa_ K123003 | 20ul | Ampicillin(50ug/ml) |
| 200ul | |||
| BBa_ K123002 | 20ul | ||
| 200ul | |||
| BBa_B0034 | 20ul | ||
| 200ul | |||
| BBa_J23100 | 20ul | ||
| 200ul | |||
| BBa_J23107 | 20ul | ||
| 200ul | |||
| BBa_R0040 | 20ul | ||
| 200ul | |||
| BBa_J23106 | 20ul | ||
| 200ul | |||
| BBa_ K398108 | 20ul | Chloramphenicol (25ug/ml) | |
| 200ul | |||
| Control | Positive (No BioBrick) | 36ul | No Antibiotic |
| Negative (No BioBrick) | 36ul | 2x Ampicillin(50ug/ml)
1x Chloramphenicol (25ug/ml) | |
Tuesday (10.7.12)
Aim - Check results from Transformation
Results: The table below indicates whether or not there was growth on each plate. Included is an image demonstrating the growth noted for BBa_K123003, BBa_K398108, and the Positive Control
| Samples | Volume Inoculated | Growth/No Growth | |
|---|---|---|---|
| BioBrick | BBa_ K123003 | 20ul | Growth |
| 200ul | Growth | ||
| BBa_ K123002 | 20ul | No Growth | |
| 200ul | No Growth | ||
| BBa_B0034 | 20ul | No Growth | |
| 200ul | No Growth | ||
| BBa_J23100 | 20ul | No Growth | |
| 200ul | No Growth | ||
| BBa_J23107 | 10ul | No Growth | |
| 2000ul | No Growth | ||
| BBa_R0040 | 20ul | No Growth | |
| 200ul | No Growth | ||
| BBa_J23106 | 20ul | No Growth | |
| 200ul | No Growth | ||
| BBa_ K398108 | 20ul | No Growth | |
| 200ul | Growth | ||
| Control | Positive (No BioBrick) | 36ul | Growth |
| Negative (No BioBrick) | 36ul | No Growth | |
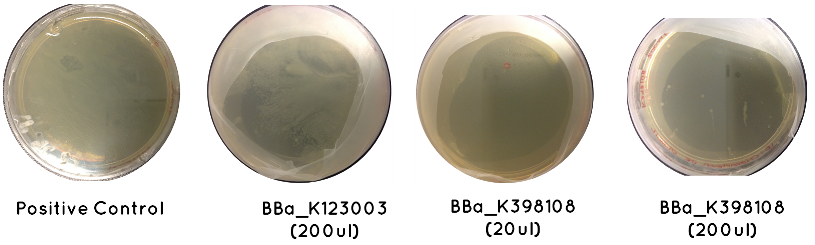
Conclusion: Only BBa_K398108 and BBa_K123003 had successfully transformed. This is a serious setback, which suggests our cell competency is not as high as we first anticipated.
Method
Step 2 - Inoculating Colonies into a Selective Broth:: Add Yul of antibiotic to reach desired antibiotic concentration.
(For Ampicillin this is 50ug/ml, For Kanamycin it is 25ug/ml, for Tetracycline it is 15ug/ml, and for Chloramphenicol it is 25ug/ml)
Step 4 – Selecting a Colony: Select a clear, isolated colony and using an inoculation hoop scoop up a colony onto the tip. Deposit in the falcon tube
Step 5 - Culture: Culture your falcon tubes overnight at a temperature of 37 oC. Leave for no longer than 16 hours.
Step 2 - Inoculating Colonies into a Selective Broth: The table below indicates the volume of broth and the concentration of antibiotic used for the BioBrick.
| Samples | Volume Inoculated | Broth (ml) | Antibiotic (ug/ml) | |
|---|---|---|---|---|
| BioBrick | BBa_ K123003 | 20ul | Lysogeny Broth (5) | Ampicillin(50ug/ml) |
| 200ul | ||||
Wednesday (11.7.12)
Aim 1 – Check Results of Picking Colonies of BBa_K398108.
Result: The broth was cloudly, indicating there has been sufficient growth of bacteria to proceed to purification (miniprep) and an Analytical Restriction Digest to determine the presence of the correct BioBrick.
Method
Step 2 - Resuspend Cells: Resuspend pelleted bacterial cells in 250ul Buffer P1 and transfer to a microcentrifuge tube
Step 3 - Puncturing Cell Membrane: Add 250ul Buffer P2 and mix thoroughly by inverting the tube 4-6 times until the solution becomes clear. Do not allow the lysis reaction to proceed for more than 5 min.
Step 4 - Neutralising buffer P2: Add 350ul Buffer N3 and mix immediately and thoroughly by inverting the tube 4-6 times.
Step 5 - Centrifuge:
RPM: 13000
Time:10 minutes
Temperature: 18oC
Step 6 - Centrifuge: Apply the supernatant from step 5 to the QIAprep spin column by decanting or pipetting. Centrifuge for 30-60s and discard the flow-through.
Step 7 - Remove Endonucleases from Sample: Wash the QIAprep spin column by adding 500ul of Buffer PB. Centrifuge for 30-60s and discard flow-through.
Step 8 - Remove salts from sample: Wash the QIAprep spin column by adding 750ul of Buffer PE. Centrifuge for 3-60s and discard flow through.
Step 9 - Centrifuge:
RPM: 13000
Time:1 minute
Temperature: 18oC
Step 10 - Elute DNA: Place the QIAprep column in a clean 1.5ml microcentrifuge tube. To elute DNA, add 50ul Buffer EB to the centre of the spin column, let it stand for 1 min, and centrifuge for 1 min.
Step 1 - Thawing cells: Thaw all materials on ice
Step 2 - Adding Ingredient: Add the following ingredients to autoclaved/sterile eppendorf tubes
| Component | Amount (ul) (one enzyme used) | Amount (ul) (two enzymes used) |
|---|---|---|
| dH20 | 2.5 | 1.5 |
| Buffer 1x | 1 | 1 |
| DNA template | 5 | 5 |
| BSA | 0.5 | 0.5 |
| Enzyme 1 | 1 | 2 |
| Enzyme 2 | N/A | 1 |
Step 3 - Addition of BioBrick: Flick contents gently and centrifuge.
Step 4 - Centrifuge:
RPM: 14000
Time: 1 minute
Temperature: 18oC
Step 5 - Digest Program: Place the samples on a thermocycler under the following conditions:
RPM: 550
Time: 2.5 hours
Temperature: 37oC
Step 6 - Denaturing Enzymes: If you are not running the samples on a gel immediately, denature the restriction enzymes by running the samples on a thermocycler under the following conditions:
RPM: 550
Time: 25 minutes
Temperature: 65oC
Step 2 – Setting up Digests and Controls: The protocol describes the recipe for (i) Digested Plasmid and (ii) Uncut Control. The table below indicates that an uncut and an Spe1/Xba1 digested sample be set up for each BioBrick.
| Samples | Recipe | Enzyme Used | Buffer | |
|---|---|---|---|---|
| BioBrick | BBa_K398108 | Digested Plasmid | Spe1 and Xba1 | 4 |
| Undigested Plasmid | None | |||
Preparing the Gel
Step 1: Within a conical flask, add 3ml 50X TAE, 1.5g Agarose, and 150ml RO water
Step 2: Heat for 1 min in microwave. Swirl. Heat again for 30s. If solution is clear stop. Else repeat.
Step 3: Cool solution under running cold water.
Step 4: Add 20ul ethidium bromide (normal concentration of EB solution is 500ug/ul)
Step 5: Pour into a sealed casting tray in a slow steady stream, ensuring there are no bubbles
Running a gel
Step 6: Add 1 part loading buffer to five parts of loading sample
Step 7: Position the gel in the tank and add TAE buffer, enough to cover the gel by several mm
Step 8: Add 5ul of DNA ladder to lane 1
Step 9: Add samples to the remaining wells
Step 10: Run at 100 volts for 1hour and 15 minutes
Imaging the Gel
Step 11: Place gel in GelDoc 2000 chamber
Step 12: Turn GelDoc 2000 chamber on
Step 13: From computer: Quantity One > Scanner > Gel_Doc_Xr>Manuqal Acquire
Step 14: Alter the exposure/settings to give a clear image.
TAE - Tris-acetate-EDTA
EDTA - ethylenediamine tetraacetic acid
Results: In Lane 1 and 2 we expect a product equivalent to the size of the pSB1C3 plasmid backbone and BBa_K398108 insert (2914bp) long, indicated by the position of A. The absence of this band, and the presence of other bands indicates that this transformation was unsuccessful. In Lane 3 and 4 we would expect a product for the BBa_K398108 insert (844bp) as indicated by B, and a product for the pSB1C3 plasmid backbone (2070bp) as indicated by C. Both products are absent, further supporting the failure of the transformation. In Lane 5 and 6 we would expect to see a similar product to Lanes 1 and 2, except for any secondary effect that the conformation of the uncut plasmid may have on its migration. Such a product was not found.
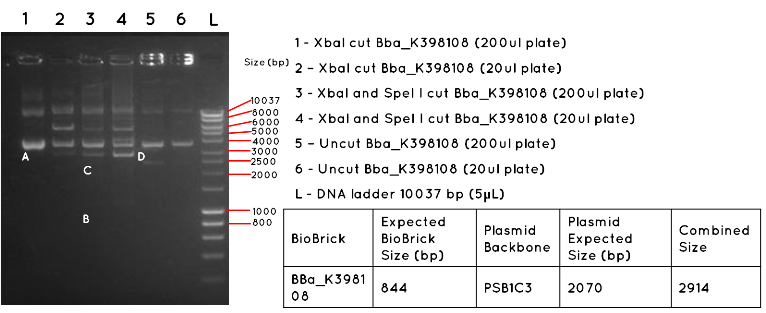
Conclusion: This transformation failed, or contamination has occurred since the colony picking.
Software ND-1000 Model:
Step 1: Initialise the spectrophotometer by pipetting 1 µ of clean water onto lower optic surface, lowering the lever arm and selecting ‘initialise’ in the ND-1000 software
Step 2: Wipe and add elution buffer as negative control. Click blank in ND-1000 software
Step 3: Wipe and add 1 µl sample
Step 4: On the software set lambda to 260nm
Step 5: Lower the lever arm and click measure in ND-1000 software
Step 6: Take readings for concentration and purity
Step 7: Once measurement complete, wipe surface
| BioBrick | λ260 | λ280 |
|---|---|---|
| BBa_K398108 | 173.8 | 186.6 |
Aim 2 - Picking Colonies for BBa_K123003. With the exception of BBa_K398108, which has already been analysed, BBa_K123003 was the only other BioBrick to show growth in the original Expt 5.1 cohort. Colonies from the Agar Plate will be selected and cultured, with the intent of purifying the plasmid for detecting (Restriction Enzyme Digest and Gel Electrophoresis) the presence of the correct BioBrick.
Method
Step 2 - Inoculating Colonies into a Selective Broth:: Add Yul of antibiotic to reach desired antibiotic concentration.
(For Ampicillin this is 50ug/ml, For Kanamycin it is 25ug/ml, for Tetracycline it is 15ug/ml, and for Chloramphenicol it is 25ug/ml)
Step 4 – Selecting a Colony: Select a clear, isolated colony and using an inoculation hoop scoop up a colony onto the tip. Deposit in the falcon tube
Step 5 - Culture: Culture your falcon tubes overnight at a temperature of 37 oC. Leave for no longer than 16 hours.
Step 2 - Inoculating Colonies into a Selective Broth: The table below indicates the volume of broth and the concentration of antibiotic used for inoculating the BioBrick.
| Samples | Volume Inoculated | Broth (ml) | Antibiotic (ug/ml) | |
|---|---|---|---|---|
| BioBrick | BBa_K123003 | 20ul | Lysogeny Broth (5) | Ampicillin(50ug/ml) |
| 200ul | ||||
Thursday 12.7.12
Aim 1 - Check results of picking colonies of BBa_K123003.
Results: There was no growth for the BioBrick.
Conclusion: The reasons for the lack of growth are not clear. As successfully transformed bacteria have already been selected for my the antibiotic positive agar plate, we would expect any colony picked to survive culture in broth of the same antibiotic concentration. It is possible that the colony did not make it into the Falcon but this is unlikely. We will repeat the picking of the BBa_K123003 colony.
Aim 2 – Picking Colonies of BBa_K123003: After we found the colony picking failed, we decided to repeat the protocol. This time however, we would make two inoculated falcons for each plate, to increase our chance of a successful culture.
Method:
Step 2 - Inoculating Colonies into a Selective Broth:: Add Yul of antibiotic to reach desired antibiotic concentration.
(For Ampicillin this is 50ug/ml, For Kanamycin it is 25ug/ml, for Tetracycline it is 15ug/ml, and for Chloramphenicol it is 25ug/ml)
Step 4 – Selecting a Colony: Select a clear, isolated colony and using an inoculation hoop scoop up a colony onto the tip. Deposit in the falcon tube
Step 5 - Culture: Culture your falcon tubes overnight at a temperature of 37 oC. Leave for no longer than 16 hours.
Step 2 - Inoculating Colonies into a Selective Broth: The table below indicates the volume of broth and the concentration of antibiotic used for inoculating the BioBrick.
| Samples | Volume Inoculated | Broth (ml) | Antibiotic (ug/ml) | |
|---|---|---|---|---|
| BioBrick | BBa_K123003 | 2x 20ul | Lysogeny Broth (5) | Ampicillin(50ug/ml) |
| 2x 200ul | ||||
Friday (13.7.12)
Aim 1 - Check results of Picking Colonies of BBa_K123003 (second attempt)
Results: There was growth for one falcon of BBa_K123003
Conclusion: Clearly BBa_K123003 is proving difficult to culture, but the presence of a single populated Falcon means we can proceed with purification (Miniprep).

Monday (9.7.12)
Aim – Overnight Culture of Original line of W3110 cells: For the Reinvigoration we will be reapplying the protocol for Generating Competent Cells on the failed cell line. To do so, we must first inoculate the original cells into a falcon, and culture overnight.
Tuesday (10.7.12)
Aim – Plating Cells on Minimal Agar: In order to culture our BioBricks in a cell line, we must first enable our cells to uptake plasmid. Today we will be starting the first day of the protocol, whereby we streak the cells on a minimal agar.
Method
Step 1 – Setting up Plates: Pour several plates with minimal agar.
Step 2 – Streaking: Streak the chosen cell line onto the minimal agar plate.
Step 3 – Incubation: Incubate at 37oC overnight
Wednesday (11.7.12)
Aim – Picking Colonies: Today we check the colony formation from yesterday’s culture, and pick a colony for inoculation
Step 1 -Creating culture media: In a sterile environment, set up a falcons, with 5mls of Lysogeny Broth and 100ul 1M MgS04.
Step 2 – Selecting a Colony: Select a clear, isolated colony and using an inoculation hoop, scoop up a colony onto the tip. Deposit in the falcon tube
Step 3 - Cell culture: Culture your falcon tubes overnight at a temperature of 37 oC. Leave for no longer than 16 hours.
Thursday (11.7.12)
Aim – Permeabilisation: Incubating Cells in the presences of CaCl2 will prepare the cell wall, such that it becomes permeable to DNA.
Step 1 – Innoculation: Inoculate 100mls of Lysogeny Broth in pre-warmed conical with 1ml of the overnight culture from Day 2
Step 2 – Incubation: Incubate for two hours in a 37oC shaker until the cells are at the early log phase of the growth curve. Measure absorbance on a spectrometer until A6000 is approximately 0.3.
Step 3 – Incubation: Transfer to a chilled sterile 50ml Falcon tube and incubate on ice for 10 minutes Step 4 – Centrifuge:
-Time: 5 mins -G – 3300 -Temperature: 180C
Step 5 – Incubate: Resus in 10ml of ice cold 0.1M CaCl2 in 15% glycerol and incubate on ice for 30 minutes.
Step 6 – Centrifuge: -Time: 5 mins -G – 3300 -Temperature: 180C
Step 7 – Transfer: Resus in 1ml of ice cold 0.1M CaCl2 in 15% glycerol. Transfer 100ul aliquots to pre-chilled, pre-labelled eppendorf tubes. Store at -70oC
Step 8 – Centrifuge -Time: 5 mins -G – 3300 -Temperature: 180C
Step 9 - Storage: Store cells at -80oC

Friday 13.7.12
Aim - Testing competency of Original W3110 Cell line: After the failure to transform the large majority of the cohort of Expt 5.1, and subsequent difficulties with culturing BBa_K123003, we are considering the possibility our W3110 cell line is not adequately competent. We are currently in the process of attempting a ‘reinvigoration’ of this cell line, but decided we should also do a formal test of the competency of the original W3110 cell line. To do so we will attempt to transform our W3110 cells with a plasmid (pTop) available from the lab, with a known concentration of 297ng/ul. At such a high concentration, transformation should be relatively easy.
If the transformation is successful, it could be the concentration of certain BioBricks (which is unknown) that is affecting the ability of the W3110s to undergo transformation. However, if transformation with pTop is not successful, it would suggest our cells are not competent.
Method
Step 1 - Thawing Cells: Thaw competent cells on ice.
Step 2 - Adding cells: Add 50 µL of thawed competent cells into pre-chilled 2ml tube.
Step 3 - Addition of BioBrick: Add 1 - 2 µL of the resuspended DNA to the 2ml tube. Pipette up and down a few times, gently. Make sure to keep the competent cells on ice.
Step 4 - Incubation: Close tube and incubate the cells on ice for 30 minutes.
Step 5 - Heat Shock: Heat shock the cells by immersion in a pre-heated water bath at 42ºC for 60 seconds.
Step 6 - Incubation: Incubate the cells on ice for 5 minutes.
Step 7 - Add media: Add 200 μl of SOC media or LB broth
Step 8 - Incubation: Incubate the cells at 37ºC for 2 hours while the tubes are rotating or shaking.
Step 9 - Label plates: Label two petri dishes with LB agar and the appropriate antibiotic(s) with the part number, plasmid backbone, and antibiotic resistance. Plate 20 µl and 200 µl of the transformation onto the dishes, and spread. This helps ensure that you will be able to pick out a single colony.
Step 10 - Culture:Incubate the plate at 37ºC for 12-14 hours, making sure the agar side of the plate is up. If incubated for too long the antibiotics start to break down and un-transformed cells will begin to grow. This is especially true for ampicillin - because the resistance enzyme is excreted by the bacteria, and inactivates the antibiotic outside of the bacteria.
Step 11 - Colony Picking: You can pick a single colony, make a glycerol stock, grow up a cell culture and miniprep.
Step 1 – Thawing Cells: Use W3110 cell line created in Week 2 (Expt 2.1)
Step 3 – Addition of BioBrick: To one 2ml eppendorf, add 1ul of pTop plasmid, and to another add nothing – this will be a control.
Step 7 – Adding Broth: Lysogeny Broth was used, as SOC media was not available.
Step 8 - Incubation: The table below indicates the Ampicillin concentration of the Agar gels. Cells cultured at 30˚C over the weekend.
| Samples | Volume Inoculated | Antibiotic in Gel | |
|---|---|---|---|
| Plasmid | pTOP | 36ul | Ampicillin (50ug/ml) |
| Controls | Positive (No plasmid) | 36ul | No Antibiotic |
| Negative (No plasmid) | 36ul | Ampicillin (50ug/ml) | |
Monday 16.7.12
Aim - Check results of Transformation. On Friday 13.7.12 our original W3110 cell line was transformed with a reference plasmid of known high concentration, as a test of the competency of our cells. Previously we had experienced difficulties in transformation (Expt 5.1) and this led us to undertake a reinvigoration of the same cell line. Simultaneously, we are running this formal test of the competency of the original cell line.
Result: The table below indicates whether there was growth. Included also are images of each Agar Plate that was set up.
| Samples | Growth/No Growth | |
|---|---|---|
| Plasmid | pTOP | No Growth |
| Control | Positive (No Plasmid) | Growth |
| Negative (No Plasid) | No Growth | |

Conclusion: Our cells do not appear to be competent. This is important because it means we will have to continue the reinvigoration, which will delay the lab work
 "
"