Team:UNAM Genomics Mexico/Parts
From 2012.igem.org
(Prototype team page) |
|||
| (28 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | < | + | {{:Template:Team:UNAM_Genomics_Mexico/webhtml| content= |
| + | __NOTOC__ | ||
| + | <br /> | ||
| + | <center><h1>'''Our Parts'''</h1></center> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <table border="0" width="850" cellspacing="15" bgcolor="transparent" cellpadding="10" id="tablecontentbg"> | ||
| + | <tr> | ||
| - | < | + | <td id="contentcolumn" align="center"><p><br /> |
| - | + | <br /> | |
| - | + | <groupparts>iGEM012 UNAM_Genomics_Mexico</groupparts> | |
| - | + | <br /> | |
| - | + | <br /> | |
| - | + | </p></td> | |
| - | + | ||
| - | </ | + | |
| - | < | + | |
| - | + | ||
| - | </ | + | |
| - | </ | + | |
| - | </ | + | |
| - | < | + | </tr> |
| + | </table> | ||
| + | <br /> | ||
| + | <br /> | ||
| - | + | <center><h1>'''Characterization'''</h1></center> | |
| - | + | <br /> | |
| - | + | <br /> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K851004 <h1>'''Aminoglycoside antibiotic resistance Sm+ Spc+'''</h1>] | ||
| + | <br /> | ||
| + | <br /> | ||
| + | The aminoglycoside antibiotic resistance gene (aadA +)of pHP45Ω was originally carried on a 1.7-kb PvuII-HindIII fragment from the R100.1 plasmid. Plasmid pHP45Ω was constructed in Pierre Prentki and Henry M. Krisch, as stated in NCBI with this fragment. The addA sequence can be found in, it is confirmed to be the one contained in pHP45Ω and the complete sequence of pHP45Ω plasmid can be found in the registry references. | ||
| + | Hawaii 2008 iGEM team attempted before to construct a similar cassette but didn’t succeed. For iGEM UNAM Genomics México 2012 project, the Ω Cassette was used in the design of an OR logic gate using a recently described new type of communication system between ''Bacillus Subtilis'' cells called Nanotubes. | ||
| + | <br> | ||
| + | <br> | ||
| - | + | <center> | |
| + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> | ||
| + | <tr> | ||
| + | <td id="contentcolumnwhite" align= "center"><br />[[File:Unamgenomicsomegacassete1.png|400px]]<br /><br /><p> | ||
| + | </p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | <br/><br/> | ||
| - | |||
| - | < | + | Second Repetition <br> |
| + | |||
| + | <center> | ||
| + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> | ||
| + | <tr> | ||
| + | <td id="contentcolumnwhite" align= "center"><br />[[File:Unamgenomicsomegacassete2.png|400px]]<br /><br /><p> | ||
| + | </p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | <br/><br/> | ||
| + | |||
| + | [http://partsregistry.org/wiki/index.php?title=Part:BBa_K851002 <h1>'''pBAD/pXyl promoter'''</h1>]<br/> | ||
| + | <br/> | ||
| + | The characterization of this construction was made on an E.coli strain in a semi quantitative attempt. In that way, we expected that only araC was repressing the promoter because the xylR binding sites corresponds to xylR of B. subtilis. Since it requires sugars for its expression, and the system is repressed by a metabolite from the pentose metabolism pathway, we used a minimal medium, M9 (Sambrook, 1989), but we changed the glucose for arginine as carbon source in order to get the lowest interference in the expression of GFP from another monosaccharides species in the medium. We use a gradient for both, xylose and arabinose on cultures at 0.1 O.D (540 nm). We could see that the expression of GFP increases as the amount of sugars added also increases. An amount of 0.01% (g/ml) of arabinose is enough for an increase of 4 times the basal expression, and the maximum production is approximately 5 times greater for the downstream genes with 0.1%(g/ml) of arabinose. Xylose gradient had a small contribution on the expression of GFP, which could be attributed to the partial similarity of the binding sites between xylR from B. subtilis and E. coli. The measurements were made with a filter fluorometer based in three different measures for each condition.<br/> | ||
| + | <br/> | ||
| + | <center> | ||
| + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> | ||
| + | <tr> | ||
| + | <td id="contentcolumnwhite" align= "center"><br />[[File:UGM Pbadxyl.png|600px]]<br /><br /><p>Expression of GFP in the sugars gradient measured in fluorescence units | ||
| + | </p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | <br/><br/> | ||
| + | |||
| + | |||
| + | }} | ||
Latest revision as of 02:29, 27 October 2012

Our Parts
|
Characterization
Aminoglycoside antibiotic resistance Sm+ Spc+
]
The aminoglycoside antibiotic resistance gene (aadA +)of pHP45Ω was originally carried on a 1.7-kb PvuII-HindIII fragment from the R100.1 plasmid. Plasmid pHP45Ω was constructed in Pierre Prentki and Henry M. Krisch, as stated in NCBI with this fragment. The addA sequence can be found in, it is confirmed to be the one contained in pHP45Ω and the complete sequence of pHP45Ω plasmid can be found in the registry references.
Hawaii 2008 iGEM team attempted before to construct a similar cassette but didn’t succeed. For iGEM UNAM Genomics México 2012 project, the Ω Cassette was used in the design of an OR logic gate using a recently described new type of communication system between Bacillus Subtilis cells called Nanotubes.
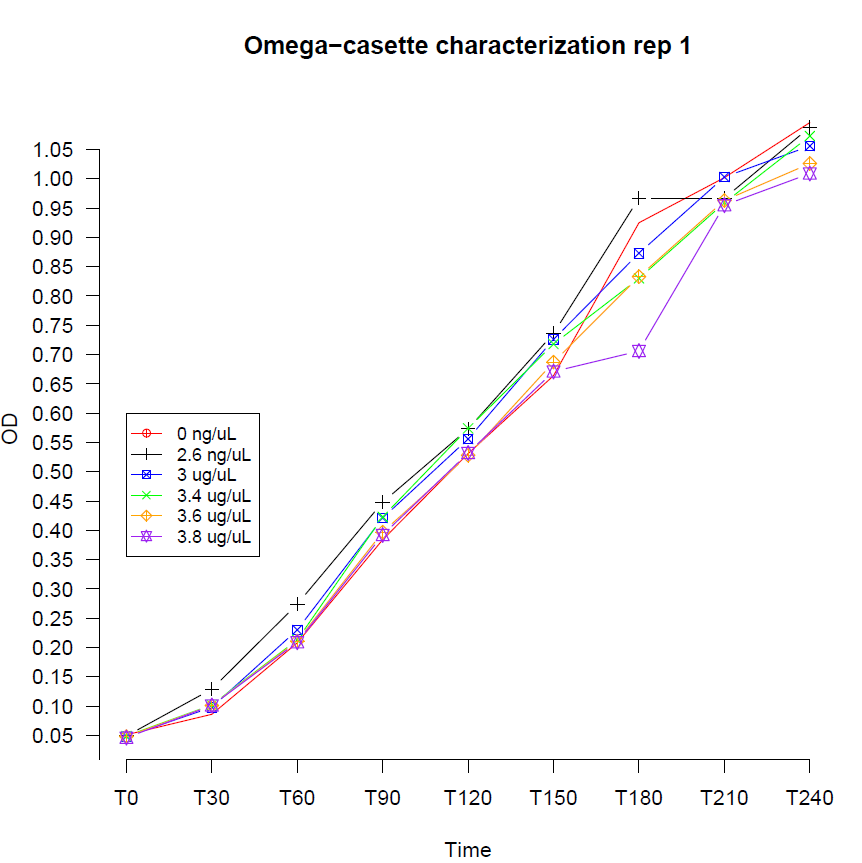
|
Second Repetition

|
pBAD/pXyl promoter
]
The characterization of this construction was made on an E.coli strain in a semi quantitative attempt. In that way, we expected that only araC was repressing the promoter because the xylR binding sites corresponds to xylR of B. subtilis. Since it requires sugars for its expression, and the system is repressed by a metabolite from the pentose metabolism pathway, we used a minimal medium, M9 (Sambrook, 1989), but we changed the glucose for arginine as carbon source in order to get the lowest interference in the expression of GFP from another monosaccharides species in the medium. We use a gradient for both, xylose and arabinose on cultures at 0.1 O.D (540 nm). We could see that the expression of GFP increases as the amount of sugars added also increases. An amount of 0.01% (g/ml) of arabinose is enough for an increase of 4 times the basal expression, and the maximum production is approximately 5 times greater for the downstream genes with 0.1%(g/ml) of arabinose. Xylose gradient had a small contribution on the expression of GFP, which could be attributed to the partial similarity of the binding sites between xylR from B. subtilis and E. coli. The measurements were made with a filter fluorometer based in three different measures for each condition.
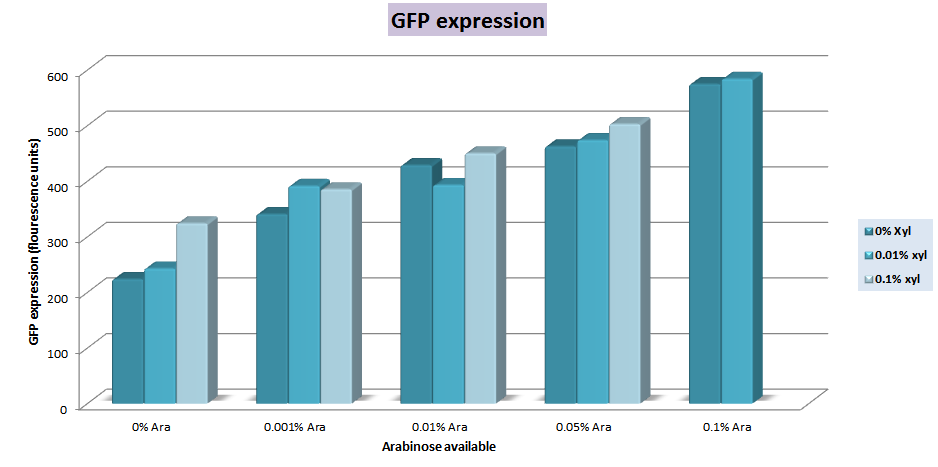 Expression of GFP in the sugars gradient measured in fluorescence units |
 "
"






