Team:UNAM Genomics Mexico/Project/DeeperDescription
From 2012.igem.org
| (42 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Template:Team:UNAM_Genomics_Mexico/webhtml| content= | {{:Template:Team:UNAM_Genomics_Mexico/webhtml| content= | ||
| + | __NOTOC__ | ||
| + | <center><h1>'''Deep Description'''</h1></center> | ||
<br /> | <br /> | ||
| - | |||
<br /> | <br /> | ||
<br /> | <br /> | ||
| + | <center> | ||
| + | <html> | ||
| + | <div class="prezi-player"><style type="text/css" media="screen">.prezi-player { width: 800px; } .prezi-player-links { text-align: center; }</style><object id="prezi_or0xn1xbzour" name="prezi_or0xn1xbzour" classid="clsid:D27CDB6E-AE6D-11cf-96B8-444553540000" width="800" height="600"><param name="movie" value="http://prezi.com/bin/preziloader.swf"/><param name="allowfullscreen" value="true"/><param name="allowFullScreenInteractive" value="true"/><param name="allowscriptaccess" value="always"/><param name="bgcolor" value="#ffffff"/><param name="flashvars" value="prezi_id=or0xn1xbzour&lock_to_path=0&color=ffffff&autoplay=no&autohide_ctrls=0"/><embed id="preziEmbed_or0xn1xbzour" name="preziEmbed_or0xn1xbzour" src="http://prezi.com/bin/preziloader.swf" type="application/x-shockwave-flash" allowfullscreen="true" allowFullScreenInteractive="true" allowscriptaccess="always" width="800" height="600" bgcolor="#ffffff" flashvars="prezi_id=or0xn1xbzour&lock_to_path=0&color=ffffff&autoplay=no&autohide_ctrls=0"></embed></object><div class="prezi-player-links"><p><a title="Copy of Project Description" href="http://prezi.com/or0xn1xbzour/copy-of-project-description/">Copy of Project Description</a> on <a href="http://prezi.com">Prezi</a></p></div></div> | ||
| + | </html> | ||
| + | </center> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | <br /> | ||
| - | + | Our boolean constructions are two, the AND & OR, for it to form a system, but how we will connect this two operations? With the nanotubes that ''Bacillus subtilis'' form. <br /> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<br /> | <br /> | ||
| - | + | ||
| + | == '''Why B. subtilis?''' == | ||
<br /> | <br /> | ||
| - | + | We found a paper in which Ben-Yehuda et. al. demostrated that ''Bacillus subtilis'' form nanotubes between them, ''Escherichia coli'' and ''Staphylococcus aureus''. And they were able to get through a GFP and some other smaller molecules. So, we decided to use this connection to form the principles of a molecular computer. How? The output of the first operation will the input for the next one, and that was what we did. Also, there is not many work in the'' B. subtilis'' at the iGEM competition, so we decided to standardize the protocols of competent cells and transformation, and to send biobricks that ''B. subtilis'' could transform and integrate for them to work correctly [[Team:UNAM_Genomics_Mexico/Notebook/Protocols#Bacillus_subtilis_Protocols | (new standard protocol)]]. <br /><br /> | |
| - | We found a paper in which Ben-Yehuda et. al. demostrated that Bacillus subtilis form nanotubes between them, Escherichia coli and Staphylococcus aureus. And they were able to get through a GFP and some other smaller molecules. So, we decided to use this connection to form the principles of a molecular computer. How? The output of the first operation will the input for the next one, and that was what we did. Also, there is not many work in the B. subtilis at the iGEM competition, so we decided to standardize the protocols of competent cells and transformation, and to send biobricks that B. subtilis could transform and integrate for them to work correctly ( | + | <center> |
<html> | <html> | ||
<div class='thumbnailWrapper'> | <div class='thumbnailWrapper'> | ||
<ul> | <ul> | ||
<li> | <li> | ||
| - | <img src='https://static.igem.org/mediawiki/2012/6/6a/Unamgenomicsdeepdescriptionbacillus1.png' height=" | + | <img src='https://static.igem.org/mediawiki/2012/6/6a/Unamgenomicsdeepdescriptionbacillus1.png' height="200" /> |
<div class='captiongray'> | <div class='captiongray'> | ||
<p class='captionInside'>2011 Gyanendra P. Dubey, Sigal Ben-Yehuda. Intercellular Nanotubes Mediate Bacterial Communication. Cell, 2011; 144 (4): 590 DOI:10.1016/j.cell.2011.01.015<br /></p> | <p class='captionInside'>2011 Gyanendra P. Dubey, Sigal Ben-Yehuda. Intercellular Nanotubes Mediate Bacterial Communication. Cell, 2011; 144 (4): 590 DOI:10.1016/j.cell.2011.01.015<br /></p> | ||
| Line 31: | Line 33: | ||
</li> | </li> | ||
<li> | <li> | ||
| - | <img src='https://static.igem.org/mediawiki/2012/c/c9/Unamgenomicsdeepdescriptionbacillus2.jpg' height=" | + | <img src='https://static.igem.org/mediawiki/2012/c/c9/Unamgenomicsdeepdescriptionbacillus2.jpg' height="200" /> |
<div class='captionnaranja'> | <div class='captionnaranja'> | ||
<p class='captionInside'>2011 Gyanendra P. Dubey, Sigal Ben-Yehuda. Intercellular Nanotubes Mediate Bacterial Communication. Cell, 2011; 144 (4): 590 DOI:10.1016/j.cell.2011.01.015<br /></p> | <p class='captionInside'>2011 Gyanendra P. Dubey, Sigal Ben-Yehuda. Intercellular Nanotubes Mediate Bacterial Communication. Cell, 2011; 144 (4): 590 DOI:10.1016/j.cell.2011.01.015<br /></p> | ||
| Line 39: | Line 41: | ||
</div> | </div> | ||
</html> | </html> | ||
| + | </center> | ||
<br /> | <br /> | ||
| + | |||
<br /> | <br /> | ||
| - | |||
<br /> | <br /> | ||
| - | The AND operator in general | + | |
| + | <br /> | ||
| + | |||
| + | <br /> | ||
| + | |||
| + | <br /> | ||
| + | |||
| + | <br /> | ||
| + | |||
| + | <br /> | ||
| + | |||
| + | <br /> | ||
| + | |||
| + | == '''How does the AND work?''' == | ||
| + | <br /> | ||
| + | <br /> | ||
| + | The AND operator in general only gives an output when the two inputs recognized in the operator are present. In the design, we considered the possibility that the constitutive promoter could continue with transcription even after getting to the double terminator and to avoid this, we designed the AND construction with the inducible promoter before the constitutive one. <br /> | ||
We have two types of AND: <br /> | We have two types of AND: <br /> | ||
| - | + | •'''The Heavy Metals AND''': The promoters CzrA/ArsR have something in common, they both sense cadmium. When the two promoters detect cadmium, they would not repress anymore and start transcription of P4 or LasR, and we will know it is working because our bacterium will be red, because of our reporter gene (RFP). The promoters are based in the CadA promoter plus a binding site for ArsR for get a response that depend in both, CzrA and ArsR. <br /> | |
| + | |||
| + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10" align="right"> | ||
| + | <tr> | ||
| + | <td id="contentcolumnwhite" align= "center"><br />[[File:UnamgenomicsCadmiopromotores.png | 430px ]]<br /><br /><p>Our CzrA/ArsR </p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | |||
| + | •'''CzrA/ArsR''': This transcription factors are involved in metal homeostasis, each one had affinity for a set of metals[1], the intersection of both sensors is Cadmium[2][3], in this case the proposed input metals will be Zinc for CzrA and Arsenic for ArsR. This TFs acts as repressors in the absence of metals, when there is metal in the cell, the metal binds to the transcription factor and make a conformation change that dissolves the affinity of the protein for their binding sites and let the promoter free to recruit RNA polymerase to start transcription[1]. <br /> | ||
| + | |||
| + | We have three''' CzrA/ArsR promoters''', one of them was designed by Newcastle team in 2009 ([http://partsregistry.org/Part:BBa_K174015 Part BBa_K174015]), the two remaining were design by our team based on the previous by Newcastle. Why we need more than one promoter? The part BBa_K174015 has no reported experience, so it could work as it should or not and the way this cadmium sensor was developed allow us to play with the combinatory of binding sites and with the -10 and -35 boxes. The cadmium sensor of Newcastle is -35, -10 and binding sites, our promoters are:<br /> | ||
| + | |||
| + | '''Promoter 1''': attta, -35,-10, ArsR binding site overlapping -35, CsrA binding site overlapping TSS, rbs+atg.<br /> | ||
| + | |||
| + | '''Promoter 2''': -35, -10, ArsR binding site overlapping after TSS, CzrA overlapping TSS, rbs+atg.<br /> | ||
| + | |||
| + | In this way, by designing different versions of the “same” part we can reach the most optimal biobrick.<br /> | ||
| + | |||
| + | •'''P4''': It´s a transcription factor from ''phi29 phage'', which is a phage of ''B. subtilis'', it is the activator of the A3 promoter[4,5]. <br /> | ||
| + | |||
| + | •'''LasR''': It is another activator transcription factor, its origin is ''Pseudomonas aeruginosa'' PAO1, its correspondent promoter is LasB[20]. <br /> | ||
| + | |||
| + | •'''CI''': This protein is a repressor from Lambda phage. <br /> | ||
| + | |||
| + | •'''RFP''': highly engineered mutant of red fluorescent protein from ''Discosoma striata'' (coral), this is our reporter gene[6]. <br /> | ||
| + | |||
| + | •'''Omega cassette''': The Omega Cassette provides resistance to Spectinomycin and Streptomycin; it was obtained from plasmid pHP45omega[7,8]. Specifically, the resistance is provided by the aminoglycoside antibiotic resistance gene (aadA +) originally carried on a 1.7-kb PvuII-HindIII fragment from the R100.1 plasmid[8]. <br /> | ||
| + | |||
| + | •'''AmyE''': Integration sequences allow DNA to be incorporated into the chromosome of a host cell at a specific locus. This is achieved by using leading (5') and trailing (3') DNA sequences that are the same as those at a specific locus of the chromosome. The 5' integration sequence([http://partsregistry.org/Part:BBa_K143001 BBa_K143001]) can be added to the front of a BioBrick construct and the 3' integration sequence specific for this locus ([http://partsregistry.org/Part:BBa_K143002 BBa_K143002]) to the rear of the Biobrick construct to allow integration of the BioBrick construct into the chromosome of the Gram-positive bacterium B. subtilis at the amyE locus[18,19]. <br /> | ||
| + | <br /> | ||
| + | <center> | ||
| + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> | ||
| + | <tr> | ||
| + | <td id="rightcolumn2" align= "center"><br />[[File:UnamgenomicsConstruccion1.png| x115px |link=Team:UNAM_Genomics_Mexico/Results/AND#CzrA-ArsR_AND_GATE]]<p>'''Heavy metal AND Results'''</p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | |||
| + | •'''The pBAD/pXyl AND''': The promoter pBAD is activated with arabinose, and pXyl with xylose. When both the promoters are activated it starts the transcription of P4 or LasR, and we will know it is working because our bacteria will be red, because of the reporter gene (RFP). <br /> | ||
| + | |||
| + | •'''pBAD''': It´s the promoter under the sigma factor 70 from ''E. coli'', which is analogous to the sigma A factor of B. subtilis. It’s repressed by AraC and induced by arabinose, which triggers the rearrangement of the AraC monomers binding for the recruitment of RNA polymerase[9,10]. <br /> | ||
| - | + | •'''pXyl''': It´s a promoter in B. subtilis which is regulated by Xylose and XylR[11,12]. In the endogenous systems, pXyl is the promoter for a set of genes involved in xylose metabolism[12]. <br /> | |
•P4: It´s a transcription factor from phi29 phage, which is a phage of B. subtilis, it is the activator of the A3 promoter[4,5]. <br /> | •P4: It´s a transcription factor from phi29 phage, which is a phage of B. subtilis, it is the activator of the A3 promoter[4,5]. <br /> | ||
| - | + | •'''LasR''': It is another activator transcription factor, its origin is ''Pseudomonas aeruginosa'' PAO1, its correspondent promoter is LasB[20]. <br /> | |
| - | + | •'''CI''': This protein is a repressor from Lambda phage. <br /> | |
| - | + | •'''RFP''': highly engineered mutant of red fluorescent protein from ''Discosoma striata'' (coral), this is our reporter gene[6]. <br /> | |
| - | + | •'''Pveg''': This is a constitutive promoter from ''B. subtilis'' regulates by the sigma factor sigma A[13]. <br /> | |
| - | + | •'''XylR''': This is a repressor transcription factor which in the presence of xylose is inactivated by a conformational change triggered by the binding with xylose that induces the dissociation of the XylR from its binding site[14]. <br /> | |
| + | |||
| + | •'''AraC''': It´s one ''E.coli'' repressor transcription factor. In the absence of arabinose, araC binds to two binding sites located in the pBAD promoter and through the binding of both monomers od AraC a DNA looping is made which blocks the transcription of the genes under that promoter[15]. When arabinose is present in the medium. <br /> | ||
| + | |||
| + | •'''Omega cassette''': The Omega Cassette provides resistance to Spectinomycin and Streptomycin; it was obtained from plasmid pHP45omega[7,8]. Specifically, the resistance is provided by the aminoglycoside antibiotic resistance gene (aadA +) originally carried on a 1.7-kb PvuII-HindIII fragment from the R100.1 plasmid[8]. <br /> | ||
| + | |||
| + | •'''AmyE''': Integration sequences allow DNA to be incorporated into the chromosome of a host cell at a specific locus. This is achieved by using leading (5') and trailing (3') DNA sequences that are the same as those at a specific locus of the chromosome. The 5' integration sequence([http://partsregistry.org/Part:BBa_K143001BBa_K143001]) can be added to the front of a BioBrick construct and the 3' integration sequence specific for this locus ([http://partsregistry.org/Part:BBa_K143002 BBa_K143002]) to the rear of the Biobrick construct to allow integration of the BioBrick construct into the chromosome of the Gram-positive bacterium B. subtilis at the amyE locus[18,19]. <br /> | ||
| + | |||
| + | <center> | ||
| + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> | ||
| + | <tr> | ||
| + | <td id="leftcolumn3" align= "center"><br />[[File:Unamgenomicsdeepconstruccion 2.png| x115px |link=Team:UNAM_Genomics_Mexico/Results/AND#ARABINOSE-XYLOSE_AND_GATE]]<p>'''Sugar AND Results'''</p></td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | |||
| + | <h2>'''How does the OR work?'''</h2><br /> | ||
| + | •The OR operator gives an output when one, or both, of the inputs recognized by the operator are present. Our OR works with the A3 and LasB promoter, that need the P4 and LasR TF, respectively. When they are active, they transcribe GFP, and we can notice it is working because of the fluorescence that GFP produces.. <br /> | ||
| + | |||
| + | •'''LasB:''' Promoter regulated positively by LasR in ''P. aeruginosa''[21]. <br /> | ||
| + | |||
| + | •'''A3''': Promoter regulated positively by P4 from phi29 phage[4,16,17]. <br /> | ||
| + | |||
| + | •'''GFP:''' The GFP reporter gene encodes green fluorescent protein derived from jellyfish ''Aequeora victoria'' wild-type GFP[22]. <br /> | ||
| + | |||
| + | •'''Omega cassette:''' The Omega Cassette provides resistance to Spectinomycin and Streptomycin; it was obtained from plasmid pHP45omega[7,8]. Specifically, the resistance is provided by the aminoglycoside antibiotic resistance gene (aadA +) originally carried on a 1.7-kb PvuII-HindIII fragment from the R100.1 plasmid[8]. <br /> | ||
| + | |||
| + | •'''AmyE:''' Integration sequences allow DNA to be incorporated into the chromosome of a host cell at a specific locus. This is achieved by using leading (5') and trailing (3') DNA sequences that are the same as those at a specific locus of the chromosome. The 5' integration sequence([http://partsregistry.org/Part:BBa_K143001 BBa_K143001]) can be added to the front of a BioBrick construct and the 3' integration sequence specific for this locus ([http://partsregistry.org/Part:BBa_K143001 BBa_K143002]) to the rear of the Biobrick construct to allow integration of the BioBrick construct into the chromosome of the Gram-positive bacterium B. subtilis at the amyE locus [18,19]. <br /> | ||
| + | <br /> | ||
| + | |||
| + | <br /> | ||
| + | |||
| + | <center> | ||
| + | <table border="0" height="150" cellspacing="15" bgcolor="transparent" id="tablecontentbg" cellpadding="10"> | ||
| + | <tr> | ||
| + | <td id="contentcolumn" align= "center"><br />[[File:Unamgenomicsconstruccion3.png | 850px|link=Team:UNAM_Genomics_Mexico/Results/OR]]</td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | |||
| + | <br /> | ||
| + | <br /> | ||
| + | ::::::::::::::::::::::::::::::::::::::[[File:UnamgenomcisUp.png|right | 120px |link=Team:UNAM_Genomics_Mexico/Project/DeeperDescription#Deep_Description]] | ||
| + | <center>[[File:Unamgenomicsdeepamye.jpg]]</center> | ||
| + | <br /> | ||
| + | <br /> | ||
| + | [1] Busenlehner LS, Pennella MA, Giedroc DP (2003). The SmtB/ArsR family of metalloregulatory transcriptional repressors: Structural insights into prokaryotic metal resistance. FEMS Microbiol Rev , 27:131-143. <br /> | ||
| + | [2] Charles M Moore and John D Helmann(2005). Metal ion homeostasis in ''Bacillus subtilis''. Current Opinion in Microbiology, 8:188–195. <br /> | ||
| + | [3] Moore CM, Gaballa A, Hui M, Ye RW, Helmann JD (2005). Genetic and physiological responses of ''Bacillus subtilis'' to metal ion stress. Mol Microbiol(1) , 27–40. <br /> | ||
| + | [4] Camacho A & Salas M (2010) DNA bending and looping in the transcriptional control of bacteriophage ϕ29. FEMS Microbiol Rev. 34(5):828-841. <br /> | ||
| + | [5] Rojo F, Mencía M, Monsalve M & Salas M (1998) Transcription activation and repression by interaction of a regulator with the a subunit of RNA polymerase: the model of phage ϕ29 protein p4. Nucleic Acid Re 60: 29–46<br /> | ||
| + | [6] http://partsregistry.org/Part:BBa_E1010<br /> | ||
| + | [7] Pierre Prentki, Anna Bind and Andrée Epstein (1991). Plasmid vectors for selecting ISI-promoted deletions in cloned DNA: sequence analysis of the omega interposon. Gene, 103:17-23. | ||
| + | [8] Pierre Prentki and Henry M. Krisch (1984). In vitro insertional mutagenesis with a selectable DNA fragment. Gene, 29:303-313<br /> | ||
| + | [9] Lee, N. (1980) Molecular Aspects of ara Regulation. In The Operon, J. H. Miller and W. S. Reznikoff, eds. (Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory), pp. 389-410. <br /> | ||
| + | [10] Lee, N., Francklyn, C., and Hamilton, E. P. (1987). Arabinose-Induced Binding of AraC Protein to araI2 Activates the araBAD Operon Promoter. Proc. Natl. Acad. Sci. USA 84, 8814-8818. <br /> | ||
| + | [11] Shamanna, D. K., and K. E. Sanderson. 1979. Genetics and regulation of D-xylose utilization in Salmonella typhimurium LT2. J. Bacteriol. 139:71-79. <br /> | ||
| + | [12] D Gartner, M Geissendorfer, & W Hillen(1988). Expression of the ''Bacillus subtilis'' xyl Operon Is Repressed at the Level of Transcription and Is Induced by Xylose J Bacteriol 170:7,3102-3109. | ||
| + | [13] Moran CP, Lang N, LeGrice SF, Lee G, Stephens M, Sonenshein AL, Pero J, Losick R (1982). Nucleotide sequences that signal the initiation of transcription and translation in ''Bacillus subtilis''. Mol Gen Genet; 186(3): 339-46 <br /> | ||
| + | [14] Kreuzer P, Gärtner D, Allmansberger R, Hillen W (1989). Identification and sequence analysis of the ''Bacillus subtilis'' W23 xylR gene and xyl operator. J Bacteriol. Jul;171(7):3840-5. <br /> | ||
| + | [15] Schlief, R. (2000). Regulation of the L-arabinose operon of ''Escherichia coli''. Trends in Genetics. 16(12):559-565. <br /> | ||
| + | [16] Sogo JM, Inciarte MR, Corral J, Viñuela E & Salas M(1979) RNA polymerase binding sites and transcription map of the DNA of ''Bacillus subtilis'' phage ϕ29. J Mol Biol 127: 411–436. | ||
| + | [17] Nuez B, Rojo F & SalasM(1992) Phage ϕ29 regulatory protein p4 stabilizes the binding of the RNA polymerase to the late promoter in a process involving direct protein–protein contact. P Natl Acad Sci USA 89: 11401–11405. <br /> | ||
| + | [18]Cutting, S M.; Vander-Horn, P B. Genetic analysis. In: Harwood C R, Cutting S M. , editors. Molecular biological methods for Bacillus. Chichester, England: John Wiley & Sons, Ltd.; 1990. pp. 27–74. <br /> | ||
| + | [19] http://partsregistry.org/Part:BBa_K143001 <br /> | ||
| + | [20]http://regtransbase.lbl.gov/cgi-bin/regtransbasepage=regulatorinfo&type=site&g | ||
| + | uid=76697<br /> | ||
| + | [21]Analysis of the Pseudomonas aeruginosa elastase (lasB) regulatory region. L Rust, E C Pesci and B H Iglewski, J. Bacteriol. February 1996 vol. 178 no. 4 1134-1140 <br /> | ||
| + | [22] http://partsregistry.org/wiki/index.php/Part:BBa_E0040<br /> | ||
}} | }} | ||
Latest revision as of 00:58, 27 October 2012

Deep Description
Our boolean constructions are two, the AND & OR, for it to form a system, but how we will connect this two operations? With the nanotubes that Bacillus subtilis form.
Why B. subtilis?
We found a paper in which Ben-Yehuda et. al. demostrated that Bacillus subtilis form nanotubes between them, Escherichia coli and Staphylococcus aureus. And they were able to get through a GFP and some other smaller molecules. So, we decided to use this connection to form the principles of a molecular computer. How? The output of the first operation will the input for the next one, and that was what we did. Also, there is not many work in the B. subtilis at the iGEM competition, so we decided to standardize the protocols of competent cells and transformation, and to send biobricks that B. subtilis could transform and integrate for them to work correctly (new standard protocol).
-

2011 Gyanendra P. Dubey, Sigal Ben-Yehuda. Intercellular Nanotubes Mediate Bacterial Communication. Cell, 2011; 144 (4): 590 DOI:10.1016/j.cell.2011.01.015
-

2011 Gyanendra P. Dubey, Sigal Ben-Yehuda. Intercellular Nanotubes Mediate Bacterial Communication. Cell, 2011; 144 (4): 590 DOI:10.1016/j.cell.2011.01.015
How does the AND work?
The AND operator in general only gives an output when the two inputs recognized in the operator are present. In the design, we considered the possibility that the constitutive promoter could continue with transcription even after getting to the double terminator and to avoid this, we designed the AND construction with the inducible promoter before the constitutive one.
We have two types of AND:
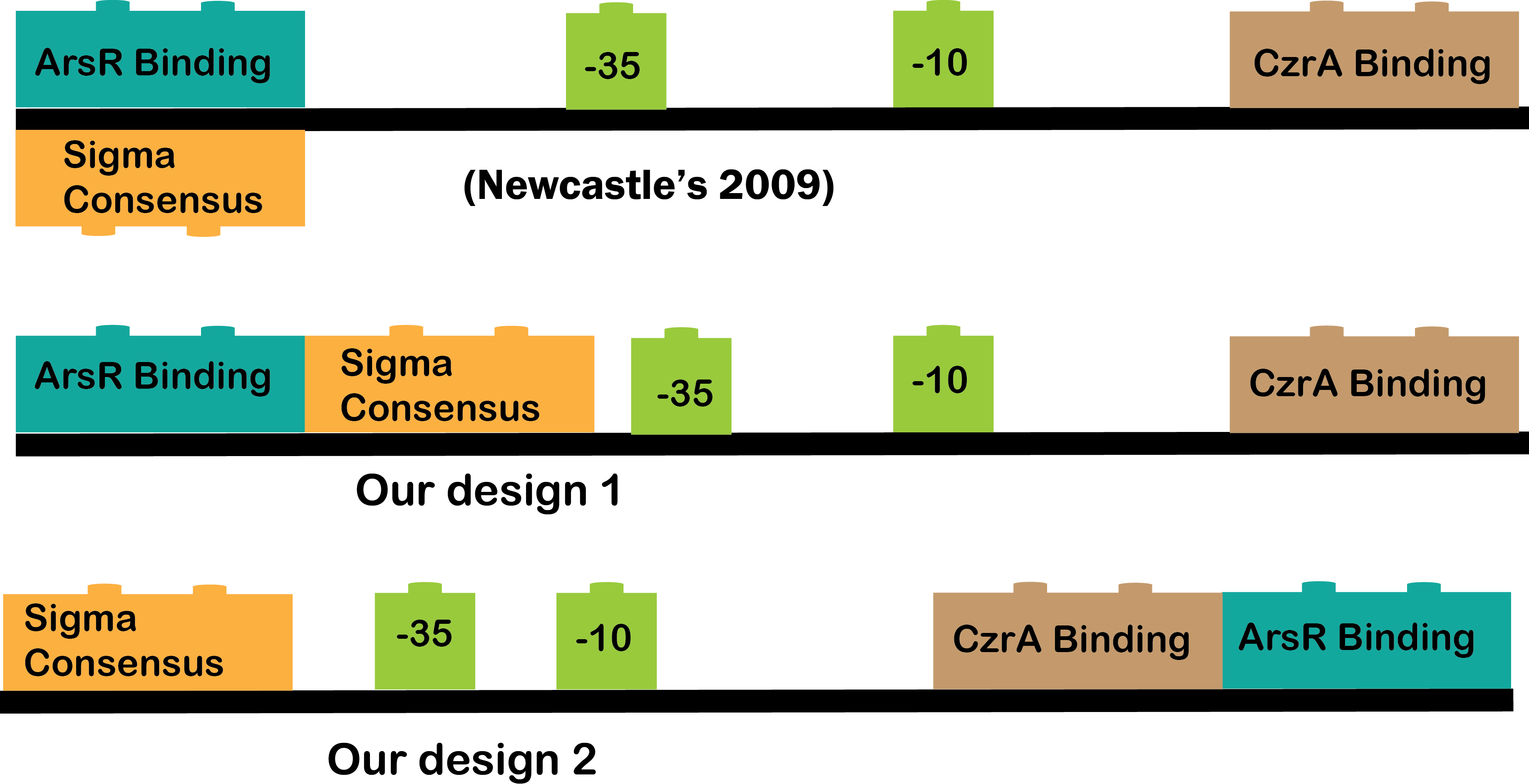
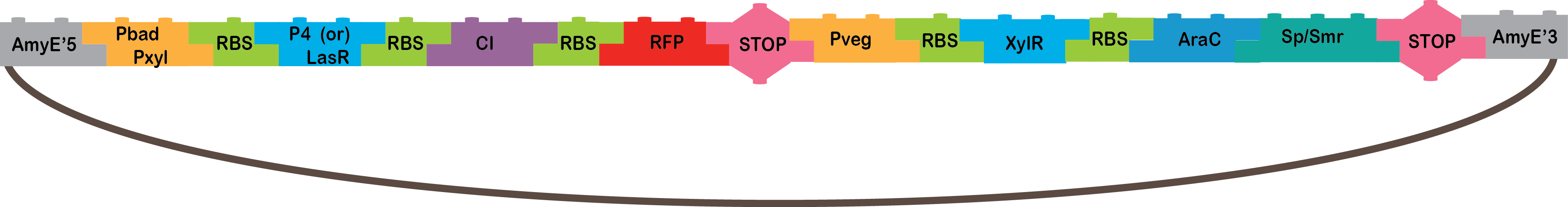
•The Heavy Metals AND: The promoters CzrA/ArsR have something in common, they both sense cadmium. When the two promoters detect cadmium, they would not repress anymore and start transcription of P4 or LasR, and we will know it is working because our bacterium will be red, because of our reporter gene (RFP). The promoters are based in the CadA promoter plus a binding site for ArsR for get a response that depend in both, CzrA and ArsR.
 Our CzrA/ArsR |
•CzrA/ArsR: This transcription factors are involved in metal homeostasis, each one had affinity for a set of metals[1], the intersection of both sensors is Cadmium[2][3], in this case the proposed input metals will be Zinc for CzrA and Arsenic for ArsR. This TFs acts as repressors in the absence of metals, when there is metal in the cell, the metal binds to the transcription factor and make a conformation change that dissolves the affinity of the protein for their binding sites and let the promoter free to recruit RNA polymerase to start transcription[1].
We have three CzrA/ArsR promoters, one of them was designed by Newcastle team in 2009 ([http://partsregistry.org/Part:BBa_K174015 Part BBa_K174015]), the two remaining were design by our team based on the previous by Newcastle. Why we need more than one promoter? The part BBa_K174015 has no reported experience, so it could work as it should or not and the way this cadmium sensor was developed allow us to play with the combinatory of binding sites and with the -10 and -35 boxes. The cadmium sensor of Newcastle is -35, -10 and binding sites, our promoters are:
Promoter 1: attta, -35,-10, ArsR binding site overlapping -35, CsrA binding site overlapping TSS, rbs+atg.
Promoter 2: -35, -10, ArsR binding site overlapping after TSS, CzrA overlapping TSS, rbs+atg.
In this way, by designing different versions of the “same” part we can reach the most optimal biobrick.
•P4: It´s a transcription factor from phi29 phage, which is a phage of B. subtilis, it is the activator of the A3 promoter[4,5].
•LasR: It is another activator transcription factor, its origin is Pseudomonas aeruginosa PAO1, its correspondent promoter is LasB[20].
•CI: This protein is a repressor from Lambda phage.
•RFP: highly engineered mutant of red fluorescent protein from Discosoma striata (coral), this is our reporter gene[6].
•Omega cassette: The Omega Cassette provides resistance to Spectinomycin and Streptomycin; it was obtained from plasmid pHP45omega[7,8]. Specifically, the resistance is provided by the aminoglycoside antibiotic resistance gene (aadA +) originally carried on a 1.7-kb PvuII-HindIII fragment from the R100.1 plasmid[8].
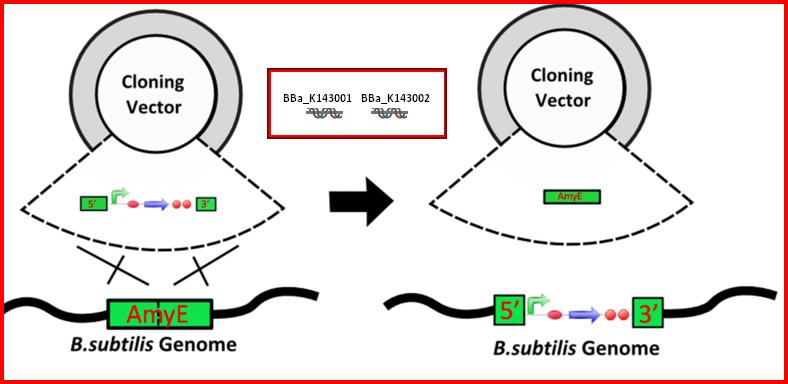
•AmyE: Integration sequences allow DNA to be incorporated into the chromosome of a host cell at a specific locus. This is achieved by using leading (5') and trailing (3') DNA sequences that are the same as those at a specific locus of the chromosome. The 5' integration sequence([http://partsregistry.org/Part:BBa_K143001 BBa_K143001]) can be added to the front of a BioBrick construct and the 3' integration sequence specific for this locus ([http://partsregistry.org/Part:BBa_K143002 BBa_K143002]) to the rear of the Biobrick construct to allow integration of the BioBrick construct into the chromosome of the Gram-positive bacterium B. subtilis at the amyE locus[18,19].
 Heavy metal AND Results |
•The pBAD/pXyl AND: The promoter pBAD is activated with arabinose, and pXyl with xylose. When both the promoters are activated it starts the transcription of P4 or LasR, and we will know it is working because our bacteria will be red, because of the reporter gene (RFP).
•pBAD: It´s the promoter under the sigma factor 70 from E. coli, which is analogous to the sigma A factor of B. subtilis. It’s repressed by AraC and induced by arabinose, which triggers the rearrangement of the AraC monomers binding for the recruitment of RNA polymerase[9,10].
•pXyl: It´s a promoter in B. subtilis which is regulated by Xylose and XylR[11,12]. In the endogenous systems, pXyl is the promoter for a set of genes involved in xylose metabolism[12].
•P4: It´s a transcription factor from phi29 phage, which is a phage of B. subtilis, it is the activator of the A3 promoter[4,5].
•LasR: It is another activator transcription factor, its origin is Pseudomonas aeruginosa PAO1, its correspondent promoter is LasB[20].
•CI: This protein is a repressor from Lambda phage.
•RFP: highly engineered mutant of red fluorescent protein from Discosoma striata (coral), this is our reporter gene[6].
•Pveg: This is a constitutive promoter from B. subtilis regulates by the sigma factor sigma A[13].
•XylR: This is a repressor transcription factor which in the presence of xylose is inactivated by a conformational change triggered by the binding with xylose that induces the dissociation of the XylR from its binding site[14].
•AraC: It´s one E.coli repressor transcription factor. In the absence of arabinose, araC binds to two binding sites located in the pBAD promoter and through the binding of both monomers od AraC a DNA looping is made which blocks the transcription of the genes under that promoter[15]. When arabinose is present in the medium.
•Omega cassette: The Omega Cassette provides resistance to Spectinomycin and Streptomycin; it was obtained from plasmid pHP45omega[7,8]. Specifically, the resistance is provided by the aminoglycoside antibiotic resistance gene (aadA +) originally carried on a 1.7-kb PvuII-HindIII fragment from the R100.1 plasmid[8].
•AmyE: Integration sequences allow DNA to be incorporated into the chromosome of a host cell at a specific locus. This is achieved by using leading (5') and trailing (3') DNA sequences that are the same as those at a specific locus of the chromosome. The 5' integration sequence([http://partsregistry.org/Part:BBa_K143001BBa_K143001]) can be added to the front of a BioBrick construct and the 3' integration sequence specific for this locus ([http://partsregistry.org/Part:BBa_K143002 BBa_K143002]) to the rear of the Biobrick construct to allow integration of the BioBrick construct into the chromosome of the Gram-positive bacterium B. subtilis at the amyE locus[18,19].
 Sugar AND Results |
How does the OR work?
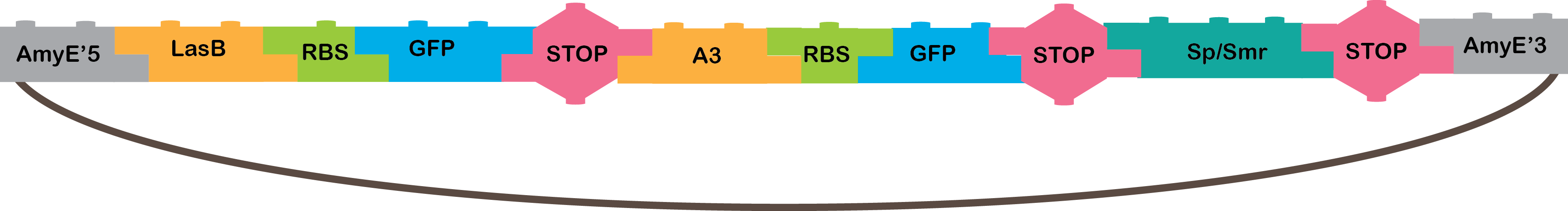
•The OR operator gives an output when one, or both, of the inputs recognized by the operator are present. Our OR works with the A3 and LasB promoter, that need the P4 and LasR TF, respectively. When they are active, they transcribe GFP, and we can notice it is working because of the fluorescence that GFP produces..
•LasB: Promoter regulated positively by LasR in P. aeruginosa[21].
•A3: Promoter regulated positively by P4 from phi29 phage[4,16,17].
•GFP: The GFP reporter gene encodes green fluorescent protein derived from jellyfish Aequeora victoria wild-type GFP[22].
•Omega cassette: The Omega Cassette provides resistance to Spectinomycin and Streptomycin; it was obtained from plasmid pHP45omega[7,8]. Specifically, the resistance is provided by the aminoglycoside antibiotic resistance gene (aadA +) originally carried on a 1.7-kb PvuII-HindIII fragment from the R100.1 plasmid[8].
•AmyE: Integration sequences allow DNA to be incorporated into the chromosome of a host cell at a specific locus. This is achieved by using leading (5') and trailing (3') DNA sequences that are the same as those at a specific locus of the chromosome. The 5' integration sequence([http://partsregistry.org/Part:BBa_K143001 BBa_K143001]) can be added to the front of a BioBrick construct and the 3' integration sequence specific for this locus ([http://partsregistry.org/Part:BBa_K143001 BBa_K143002]) to the rear of the Biobrick construct to allow integration of the BioBrick construct into the chromosome of the Gram-positive bacterium B. subtilis at the amyE locus [18,19].
 |
[1] Busenlehner LS, Pennella MA, Giedroc DP (2003). The SmtB/ArsR family of metalloregulatory transcriptional repressors: Structural insights into prokaryotic metal resistance. FEMS Microbiol Rev , 27:131-143.
[2] Charles M Moore and John D Helmann(2005). Metal ion homeostasis in Bacillus subtilis. Current Opinion in Microbiology, 8:188–195.
[3] Moore CM, Gaballa A, Hui M, Ye RW, Helmann JD (2005). Genetic and physiological responses of Bacillus subtilis to metal ion stress. Mol Microbiol(1) , 27–40.
[4] Camacho A & Salas M (2010) DNA bending and looping in the transcriptional control of bacteriophage ϕ29. FEMS Microbiol Rev. 34(5):828-841.
[5] Rojo F, Mencía M, Monsalve M & Salas M (1998) Transcription activation and repression by interaction of a regulator with the a subunit of RNA polymerase: the model of phage ϕ29 protein p4. Nucleic Acid Re 60: 29–46
[6] http://partsregistry.org/Part:BBa_E1010
[7] Pierre Prentki, Anna Bind and Andrée Epstein (1991). Plasmid vectors for selecting ISI-promoted deletions in cloned DNA: sequence analysis of the omega interposon. Gene, 103:17-23.
[8] Pierre Prentki and Henry M. Krisch (1984). In vitro insertional mutagenesis with a selectable DNA fragment. Gene, 29:303-313
[9] Lee, N. (1980) Molecular Aspects of ara Regulation. In The Operon, J. H. Miller and W. S. Reznikoff, eds. (Cold Spring Harbor, N.Y.: Cold Spring Harbor Laboratory), pp. 389-410.
[10] Lee, N., Francklyn, C., and Hamilton, E. P. (1987). Arabinose-Induced Binding of AraC Protein to araI2 Activates the araBAD Operon Promoter. Proc. Natl. Acad. Sci. USA 84, 8814-8818.
[11] Shamanna, D. K., and K. E. Sanderson. 1979. Genetics and regulation of D-xylose utilization in Salmonella typhimurium LT2. J. Bacteriol. 139:71-79.
[12] D Gartner, M Geissendorfer, & W Hillen(1988). Expression of the Bacillus subtilis xyl Operon Is Repressed at the Level of Transcription and Is Induced by Xylose J Bacteriol 170:7,3102-3109.
[13] Moran CP, Lang N, LeGrice SF, Lee G, Stephens M, Sonenshein AL, Pero J, Losick R (1982). Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet; 186(3): 339-46
[14] Kreuzer P, Gärtner D, Allmansberger R, Hillen W (1989). Identification and sequence analysis of the Bacillus subtilis W23 xylR gene and xyl operator. J Bacteriol. Jul;171(7):3840-5.
[15] Schlief, R. (2000). Regulation of the L-arabinose operon of Escherichia coli. Trends in Genetics. 16(12):559-565.
[16] Sogo JM, Inciarte MR, Corral J, Viñuela E & Salas M(1979) RNA polymerase binding sites and transcription map of the DNA of Bacillus subtilis phage ϕ29. J Mol Biol 127: 411–436.
[17] Nuez B, Rojo F & SalasM(1992) Phage ϕ29 regulatory protein p4 stabilizes the binding of the RNA polymerase to the late promoter in a process involving direct protein–protein contact. P Natl Acad Sci USA 89: 11401–11405.
[18]Cutting, S M.; Vander-Horn, P B. Genetic analysis. In: Harwood C R, Cutting S M. , editors. Molecular biological methods for Bacillus. Chichester, England: John Wiley & Sons, Ltd.; 1990. pp. 27–74.
[19] http://partsregistry.org/Part:BBa_K143001
[20]http://regtransbase.lbl.gov/cgi-bin/regtransbasepage=regulatorinfo&type=site&g
uid=76697
[21]Analysis of the Pseudomonas aeruginosa elastase (lasB) regulatory region. L Rust, E C Pesci and B H Iglewski, J. Bacteriol. February 1996 vol. 178 no. 4 1134-1140
[22] http://partsregistry.org/wiki/index.php/Part:BBa_E0040
 "
"







