Team:SUSTC-Shenzhen-A/Biosearch Progress
From 2012.igem.org
(Created page with "{{Template:SUSTC_A2}} <html> <head> <style type="text/css"> .sidebar_box_woframe { margin-bottom: 20px; } .sidebar_box { width: 260px; padding: 0; } .sidebar_box_top { wi...") |
|||
| (35 intermediate revisions not shown) | |||
| Line 160: | Line 160: | ||
<body > | <body > | ||
| - | <table width="899" border="0" cellspacing="10px" cellpadding="10px"align="center" | + | <table width="899" border="0" cellspacing="10px" cellpadding="10px"align="center" |
| - | + | <tr> | |
| - | <td valign="top"> | + | <td width="600" valign="top"> |
| - | < | + | <h1 class="title">Progress</h1> |
<p> </p> | <p> </p> | ||
| - | <p class="title1"> | + | |
| - | <p> | + | <p class="title1">App Development</p> |
| + | <p> To make sure the apps in the app store is of great quality, Apple has very strict reviewing rules for apps submitted by developers.</p> | ||
| + | <p> First,the user interface. Apple offers developers dozens of UI elements ,which is all in the Apple Style.Developers are encouraged to use Apple designing patterns so that the user can get used to a new App immediately.</p> | ||
| + | <p> For security problem as well as the efficiency, Apple requires developers to use public APIs given by Apple. This is very important,developers should be very careful about the APIs in their app.</p> | ||
| + | <p> Of course,The app can not access the user's information without asking for permission.</p> | ||
| + | <p> One of the most important features for a good app is that it should not crash. Developers should test carefully on different devices with different iOS version before they submit the app to Apple.</p> | ||
| + | <p> Last but most important. An app should give users great experience. There is so much work to do to make an app perfect because there are so many details to handle. For example, every picture needs to be optimized so that it can work on devices with retina display. Developers should also offer a user guide to make sure the users can manage to use the app very easily.</p> | ||
<p> </p> | <p> </p> | ||
| - | < | + | <p class="title1">Search Part</p> |
| - | <p> If | + | <p> To see the main interface, please refer to Fig.1. You can find your wanting BioBricks via this search part. Type the name of a BioBrick, then press the Search button on the right top. Then our software will return the BioBrick (Refer to Fig.2). If you cannot remember the full name of the BioBricks, you can submit only part of the name. For example, if you want to get the information of ‘BBa_I14050’, you type in ‘BBa_I’, the software will return the BioBricks that contain the part name ‘BBa_I’. Sometimes you may find it hard to remember the name of a BioBrick. Other search methods will help you find the target BioBrick. Submit a keyword, our software will return the BioBircks which have some relationship with this keyword (Refer to Fig.3). Moreover, you can choose the chassis a BioBrick in by press the Chassis button (Refer to Fig.4). You can choose from a variety of choices such as E.coli and yeast (Refer to Fig.5_1). If you don’t choose a specific chassis, our software will find the BioBricks from all chassis. Similarly, you can choose to search a specific type of BioBricks, e.g. terminator (Refer to Fig.5_2). </p> |
| + | <p> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/9/90/Fig.1.png" width="190"title="fig.1"> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/1/19/Fig.2.png" width="190"title="fig.2"> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/e/e3/Fig.3.png" width="190"title="fig.3"><br/> | ||
| + | Fig.1 | ||
| + | Fig.2 | ||
| + | Fig.3 | ||
| + | </p> | ||
| + | <p> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/2/2c/Fig.4.png" width="190"title="fig.4"> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/a/af/Fig.5-1.png" width="190"title="fig.5-1"> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/2/2c/Fig.5-2.png" width="190"title="fig.5-2"> | ||
| + | Fig.4 | ||
| + | Fig.5-1 | ||
| + | Fig.5-2 | ||
| + | </p> | ||
| + | <p> The default choice is to search in all types of BioBricks (Refer to Fig.6). If you find the above options cannot meet your standard, you can find some advanced options by press the edit button on top right (Refer to Fig.7). Three advanced options are available. The first is the author option. If you know the name of the creator of some BioBricks, you can refine the search by typing his/her name. If you yourself has created some BioBricks, why not try your name to see whether you can see your BioBricks? (Refer to Fig.8) The second choice is the status option. The status of a BioBrick reveals whether it is available or not. Of course, some BioBricks are also labeled ‘planning’, ‘deleted’ etc. (Refer to Fig.9) The last choice is the creation date option. If you want to pick up the BioBricks created during a period of time. Chosse a start and an end date, then our software will return the BioBricks which created during this period. (Refer to Fig.10) The first BioBrick was created on July 6th 2003. So you cannot pick a start date earlier than this.(Refer to Fig.11)</p> | ||
| + | </p> | ||
| + | <p> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/1/1a/Fig.6.png" width="190"title="fig.6"> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/2/2b/Fig.7.png" width="190"title="fig.7"> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/9/95/Fig.8.png" width="190"title="fig.8"> | ||
| + | Fig.6 | ||
| + | Fig.7 | ||
| + | Fig.8 | ||
| + | </p> | ||
| + | <p> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/9/93/Fig.9.png" width="190"title="fig.9"> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/f/f2/Fig.10.png" width="190"title="fig.10"> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/5/56/Fig.11.png" width="190"title="fig.11"> | ||
| + | Fig.9 | ||
| + | Fig.10 | ||
| + | Fig.11 | ||
| + | </p> | ||
| + | <p> Neither can you pick an end date later than the current date. (Refer to Fig.12) In addition, a start date should be earlier than an end date. So if you don’t obey this rule, our software will tell the mistake you make and reset the date. (Refer to Fig.13) There is a tutorial button lies in the top left. Click it, a short tutorial will be shown. (Refer to Fig.14)</p> | ||
| + | <p> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/4/4b/Fig.12.png" width="190"title="fig.12"> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/b/b0/Fig.13.png" width="190"title="fig.13"> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/7/7e/Fig.14.png" width="190"title="fig.14"> | ||
| + | Fig.12 | ||
| + | Fig.13 | ||
| + | Fig.14 | ||
| + | </p> | ||
| + | <p> You don’t have to make a choice on every choice. If you make no choice and press the search button, you have to spend some time waiting our software to return all the BioBricks. After you have made your own choices, you need to press the search button in the top right to see the search results. (Refer to Fig.15) Our software shows results in a list form. The name, short description and the status of a BioBrick are present in a row. (Refer to Fig.16) The search results are carefully arranged. You will find the available BioBricks always lie before the non-available (including ‘planning’, ‘deleted’, ‘unavailable’ etc.) ones. And the ones labeled with ‘in stock’ lie before the ones with ‘not in stock’. If two BioBricks are both available and in stock, the one with longer description will be shown first. Moreover if two BioBricks share the same length of description and both available and in stock (though this is very rare), the one with later creation date will be shown first. And if you find the numerous results upset you, you can press the search button in the top left to do the second search. The second search is not so complicated as the first. Only keywords are needed. Type in your wanting description, our software will find the target from the first search results. (Refer to Fig.17) If you want to return to the first search page, press the button named with ‘Search’ in the top left. (Not the search button in the top right!) (Refer to Fig.18) </p> | ||
| + | <p> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/2/22/Fig.15.png" width="190"title="fig.15"> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/f/ff/Fig.16.png" width="190"title="fig.16"> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/0/02/Fig.17.png" width="190"title="fig.17"> | ||
| + | Fig.15 | ||
| + | Fig.16 | ||
| + | Fig.17 | ||
| + | </p> | ||
| + | |||
| + | <p> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/e/ee/Fig.18.png" width="140"title="fig.18"> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/e/ec/Fig.19.png" width="140"title="fig.19"> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/c/c9/Fig.20.png" width="140"title="fig.20"> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/1/14/Fig.21.png" width="140"title="fig.21"> | ||
| + | Fig.18 | ||
| + | Fig.19 | ||
| + | Fig.20 | ||
| + | Fig.21 | ||
| + | </p> | ||
| + | <p> If you find this BioBrick terrific, you can add it to your favorites. Click the button named ‘add’ in the top right. (Refer to Fig.22) A pop-up alert view allows you to type in a nickname for this BioBrick if you like. The default nickname is ‘None’. (Refer to Fig.23) Then you can find your BioBrick in the bookmark part. (Refer to Fig.24)</p> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/a/ab/Fig.22.png" width="190"title="fig.22"> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/c/cf/Fig.23.png" width="190"title="fig.23"> | ||
| + | <img src="https://static.igem.org/mediawiki/igem.org/d/dd/Fig.24.png" width="190"title="fig.24"> | ||
| + | Fig.22 | ||
| + | Fig.23 | ||
| + | Fig.24 | ||
<p> </p> | <p> </p> | ||
| - | < | + | <p class="title1">Catalog Part</p> |
| - | <p> | + | <p> This part provides users a way to browse all the BioBricks by several different classification approaches. We studied the categories of the BioBricks provided by the Registry, and divided the BioBricks in four parts, like “parts by type”,“ devices by type”, etc. In each section, there are more precise classifications. For example, in the “parts by type” section, there are many parts like “promoter”, “RBS” and so on; users can find the BioBricks that they want in this section. |
| + | There are three parts under each section; they are “list”, “description” and “design”. List is where all the BioBricks are listed. Because sometimes the list can be very long, so there is a search function, by which users can find a specific BioBrick in the list. Description part is the general description of this type of BioBricks. Some types like promoter have a “design” part, which is a concise tutorial on how to design a BioBrick.</p> | ||
<p> </p> | <p> </p> | ||
| - | < | + | <p class="title1">Detailed Page Part</p> |
| - | <p> | + | <p><strong>Background Operation :</strong> show as below</p> |
| + | <p><img src="https://static.igem.org/mediawiki/2012/2/2b/Background_Operation.png" width="540"></p> | ||
| + | <p><img src="https://static.igem.org/mediawiki/2012/3/36/SUSTC_Original_idea.png" width="540"></p> | ||
<p> </p> | <p> </p> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</td> | </td> | ||
| - | + | <td valign="top"> | |
| + | <p class="center"><b>Below pics are ex-UIs of BioSearch</b></p> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/7/70/SUSTC_Cateloghome1.jpg" width="240"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/c/ca/SUSTC_Cateloghome2.jpg" width="240"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/6/62/Cateloghome4.jpg" width="240"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/4/4d/SUSTC_Cateloghome5.jpg" width="240"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/4/4a/SUSTC_Cateloghome3.jpg"width="240"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/5/51/SUSTC_Mainpage.jpg" width="240"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/d/d4/Screen_Shot_2012-07-26_at_5.32.14_PM.png" width="240"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/6/68/SUSTC_7.jpg" width="240"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/6/6c/Screen_Shot_2012-07-26_at_5.31.45_PM.png" width="240"> | ||
| + | <img src="https://static.igem.org/mediawiki/2012/5/50/SUSTC_5.jpg" width="240"> | ||
| + | |||
| + | </td> | ||
| + | </tr> | ||
</table> | </table> | ||
| - | |||
| - | |||
| - | |||
Latest revision as of 22:41, 26 September 2012
Progress
App Development To make sure the apps in the app store is of great quality, Apple has very strict reviewing rules for apps submitted by developers. First,the user interface. Apple offers developers dozens of UI elements ,which is all in the Apple Style.Developers are encouraged to use Apple designing patterns so that the user can get used to a new App immediately. For security problem as well as the efficiency, Apple requires developers to use public APIs given by Apple. This is very important,developers should be very careful about the APIs in their app. Of course,The app can not access the user's information without asking for permission. One of the most important features for a good app is that it should not crash. Developers should test carefully on different devices with different iOS version before they submit the app to Apple. Last but most important. An app should give users great experience. There is so much work to do to make an app perfect because there are so many details to handle. For example, every picture needs to be optimized so that it can work on devices with retina display. Developers should also offer a user guide to make sure the users can manage to use the app very easily.
Search Part To see the main interface, please refer to Fig.1. You can find your wanting BioBricks via this search part. Type the name of a BioBrick, then press the Search button on the right top. Then our software will return the BioBrick (Refer to Fig.2). If you cannot remember the full name of the BioBricks, you can submit only part of the name. For example, if you want to get the information of ‘BBa_I14050’, you type in ‘BBa_I’, the software will return the BioBricks that contain the part name ‘BBa_I’. Sometimes you may find it hard to remember the name of a BioBrick. Other search methods will help you find the target BioBrick. Submit a keyword, our software will return the BioBircks which have some relationship with this keyword (Refer to Fig.3). Moreover, you can choose the chassis a BioBrick in by press the Chassis button (Refer to Fig.4). You can choose from a variety of choices such as E.coli and yeast (Refer to Fig.5_1). If you don’t choose a specific chassis, our software will find the BioBricks from all chassis. Similarly, you can choose to search a specific type of BioBricks, e.g. terminator (Refer to Fig.5_2).
The default choice is to search in all types of BioBricks (Refer to Fig.6). If you find the above options cannot meet your standard, you can find some advanced options by press the edit button on top right (Refer to Fig.7). Three advanced options are available. The first is the author option. If you know the name of the creator of some BioBricks, you can refine the search by typing his/her name. If you yourself has created some BioBricks, why not try your name to see whether you can see your BioBricks? (Refer to Fig.8) The second choice is the status option. The status of a BioBrick reveals whether it is available or not. Of course, some BioBricks are also labeled ‘planning’, ‘deleted’ etc. (Refer to Fig.9) The last choice is the creation date option. If you want to pick up the BioBricks created during a period of time. Chosse a start and an end date, then our software will return the BioBricks which created during this period. (Refer to Fig.10) The first BioBrick was created on July 6th 2003. So you cannot pick a start date earlier than this.(Refer to Fig.11)
Neither can you pick an end date later than the current date. (Refer to Fig.12) In addition, a start date should be earlier than an end date. So if you don’t obey this rule, our software will tell the mistake you make and reset the date. (Refer to Fig.13) There is a tutorial button lies in the top left. Click it, a short tutorial will be shown. (Refer to Fig.14)
You don’t have to make a choice on every choice. If you make no choice and press the search button, you have to spend some time waiting our software to return all the BioBricks. After you have made your own choices, you need to press the search button in the top right to see the search results. (Refer to Fig.15) Our software shows results in a list form. The name, short description and the status of a BioBrick are present in a row. (Refer to Fig.16) The search results are carefully arranged. You will find the available BioBricks always lie before the non-available (including ‘planning’, ‘deleted’, ‘unavailable’ etc.) ones. And the ones labeled with ‘in stock’ lie before the ones with ‘not in stock’. If two BioBricks are both available and in stock, the one with longer description will be shown first. Moreover if two BioBricks share the same length of description and both available and in stock (though this is very rare), the one with later creation date will be shown first. And if you find the numerous results upset you, you can press the search button in the top left to do the second search. The second search is not so complicated as the first. Only keywords are needed. Type in your wanting description, our software will find the target from the first search results. (Refer to Fig.17) If you want to return to the first search page, press the button named with ‘Search’ in the top left. (Not the search button in the top right!) (Refer to Fig.18)
If you find this BioBrick terrific, you can add it to your favorites. Click the button named ‘add’ in the top right. (Refer to Fig.22) A pop-up alert view allows you to type in a nickname for this BioBrick if you like. The default nickname is ‘None’. (Refer to Fig.23) Then you can find your BioBrick in the bookmark part. (Refer to Fig.24) 

 Fig.22
Fig.23
Fig.24
Fig.22
Fig.23
Fig.24
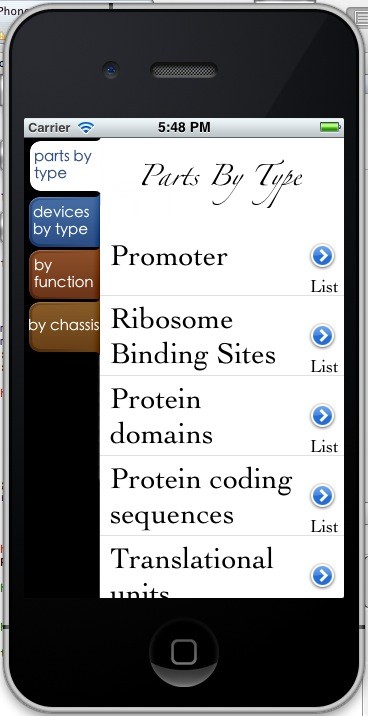
Catalog Part This part provides users a way to browse all the BioBricks by several different classification approaches. We studied the categories of the BioBricks provided by the Registry, and divided the BioBricks in four parts, like “parts by type”,“ devices by type”, etc. In each section, there are more precise classifications. For example, in the “parts by type” section, there are many parts like “promoter”, “RBS” and so on; users can find the BioBricks that they want in this section. There are three parts under each section; they are “list”, “description” and “design”. List is where all the BioBricks are listed. Because sometimes the list can be very long, so there is a search function, by which users can find a specific BioBrick in the list. Description part is the general description of this type of BioBricks. Some types like promoter have a “design” part, which is a concise tutorial on how to design a BioBrick.
Detailed Page Part Background Operation : show as below
|
Below pics are ex-UIs of BioSearch 








|
 "
"









 Fig.4
Fig.5-1
Fig.5-2
Fig.4
Fig.5-1
Fig.5-2

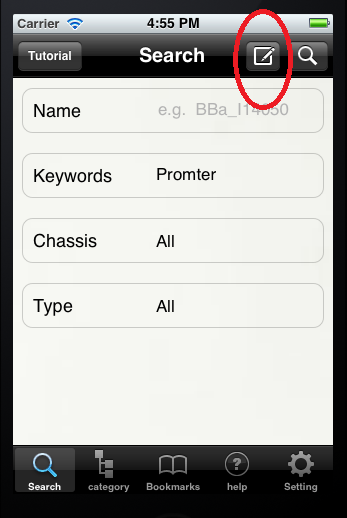
 Fig.6
Fig.7
Fig.8
Fig.6
Fig.7
Fig.8


 Fig.9
Fig.10
Fig.11
Fig.9
Fig.10
Fig.11


 Fig.12
Fig.13
Fig.14
Fig.12
Fig.13
Fig.14


 Fig.15
Fig.16
Fig.17
Fig.15
Fig.16
Fig.17



 Fig.18
Fig.19
Fig.20
Fig.21
Fig.18
Fig.19
Fig.20
Fig.21


