Team:SUSTC-Shenzhen-A/Project
From 2012.igem.org
(→Motivation) |
|||
| (74 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | {{Template:SUSTC_A2}} | |
| - | + | ||
<html> | <html> | ||
| - | < | + | <head> |
| - | < | + | <style type="text/css"> |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | .sidebar_box_woframe { | |
| + | margin-bottom: 20px; | ||
| + | } | ||
| + | .sidebar_box { | ||
| + | width: 260px; | ||
| + | padding: 0; | ||
| - | + | } | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | .sidebar_box_top { | ||
| + | width: 260px; | ||
| + | height: 20px; | ||
| + | } | ||
| + | .sidebar_box_bottom { | ||
| + | width: 260px; | ||
| + | height: 20px; | ||
| + | margin-bottom: 30px; | ||
| - | + | } | |
| - | + | ||
| - | + | .sidebar_box_content { | |
| - | + | padding: 0 20px; | |
| - | + | } | |
| - | + | ||
| - | + | .sidebar_box_content p { | |
| - | + | margin-bottom: 10px; | |
| + | } | ||
| - | + | .image_wrapper { | |
| - | + | position: relative; | |
| + | display: block; | ||
| + | width: 218px; | ||
| + | height: 128px; | ||
| + | padding: 9px; | ||
| + | } | ||
| + | .image_wrapper span { | ||
| + | position: absolute; | ||
| + | left: 0; | ||
| + | top: 0; | ||
| + | width: 236px; | ||
| + | height: 144px; | ||
| + | background: url(https://static.igem.org/mediawiki/2012/3/3d/Fruit_site_templatemo_image_frame.png) no-repeat; | ||
| + | } | ||
| + | .image_fl { float: left; margin: 3px 20px 0 0; } | ||
| + | .image_fr { float: right; margin: 3px 0 0 20px; } | ||
| - | + | .title { | |
| + | font-family: "Comic Sans MS", cursive; | ||
| + | font-size: xx-large; | ||
| + | color: #698B69; | ||
| + | } | ||
| + | .title1 { | ||
| + | font-family: "Comic Sans MS", cursive; | ||
| + | font-size: x-large; | ||
| + | color: #000; | ||
| + | } | ||
| + | .date { | ||
| + | font-family: serif, cursive, fantasy; | ||
| + | color:#5f3c23; | ||
| + | } | ||
| + | #talkbubble_a { | ||
| + | width: 544px; | ||
| + | height:330px; | ||
| + | // background:#EFEFEF; | ||
| + | background:#ffffff; | ||
| + | position: relative; | ||
| + | -moz-border-radius: 0px; | ||
| + | -webkit-border-radius: 0px; | ||
| + | border-radius: 10px; | ||
| + | font-family: Tahoma, Geneva, sans-serif; | ||
| + | font-size: 13px; | ||
| + | color:#333; | ||
| + | } | ||
| + | #talkbubble_b { | ||
| + | width: 544px; | ||
| + | height:1px; | ||
| + | background:#EFEFEF; | ||
| + | // background:#ffffff; | ||
| + | position: relative; | ||
| + | -moz-border-radius: 0px; | ||
| + | -webkit-border-radius: 0px; | ||
| + | border-radius: 10px; | ||
| + | font-family: Tahoma, Geneva, sans-serif; | ||
| + | font-size: 13px; | ||
| + | color:#333; | ||
| + | } | ||
| + | #talkbubble_c1 { | ||
| + | width: 859px; | ||
| + | height:400px; | ||
| + | background:#EFEFEF; | ||
| + | // background:#ffffff; | ||
| + | position: relative; | ||
| + | -moz-border-radius: 0px; | ||
| + | -webkit-border-radius: 0px; | ||
| + | border-radius: 10px; | ||
| + | font-family: Tahoma, Geneva, sans-serif; | ||
| + | font-size: 13px; | ||
| + | color:#333; | ||
| + | } | ||
| + | #talkbubble_c2 { | ||
| + | width: 862px; | ||
| + | height:400px; | ||
| + | // background:#EFEFEF; | ||
| + | background:#ffffff; | ||
| + | position: relative; | ||
| + | -moz-border-radius: 0px; | ||
| + | -webkit-border-radius: 0px; | ||
| + | border-radius: 10px; | ||
| + | font-family: Tahoma, Geneva, sans-serif; | ||
| + | font-size: 13px; | ||
| + | color:#333; | ||
| + | } | ||
| + | #talkbubble_c3 { | ||
| + | width: 859px; | ||
| + | height:200px; | ||
| + | background:#EFEFEF; | ||
| + | // background:#ffffff; | ||
| + | position: relative; | ||
| + | -moz-border-radius: 0px; | ||
| + | -webkit-border-radius: 0px; | ||
| + | border-radius: 10px; | ||
| + | font-family: Tahoma, Geneva, sans-serif; | ||
| + | font-size: 13px; | ||
| + | color:#333; | ||
| + | } | ||
| + | #talkbubble1 { | ||
| + | width: 275px; | ||
| + | height:330px; | ||
| + | // background:#FAF0E6; | ||
| + | background:#f2eada; | ||
| + | position: relative; | ||
| + | -moz-border-radius: 10px; | ||
| + | -webkit-border-radius: 10px; | ||
| + | border-radius: 10px; | ||
| + | text-align: left; | ||
| + | border-top-style: solid; | ||
| + | border-right-style: solid; | ||
| + | border-bottom-style: solid; | ||
| + | border-left-style: solid; | ||
| + | border-width: thin; | ||
| + | border-top-color:#DCDCDC; | ||
| + | border-right-color: #DCDCDC; | ||
| + | border-bottom-color:#DCDCDC; | ||
| + | border-left-color:#DCDCDC; | ||
| + | font-size: 14px; | ||
| + | color:#333; | ||
| + | font-family: Tahoma, Geneva; | ||
| + | } | ||
| + | </style> | ||
| + | </head> | ||
| + | <body > | ||
| + | <table width="899" border="0" cellspacing="10px" cellpadding="10px"align="center"> | ||
| + | <tr> | ||
| + | <td valign="top"> | ||
| + | <h1 class="title"><strong>Project Overview</strong></h1> | ||
| + | <p class="title1"> Biosearch</p> | ||
| + | <p><img src="https://static.igem.org/mediawiki/2012/e/e4/BiosearchPage1.png" valign="top" align="right" width="100" style="margin:10px;" /></p> | ||
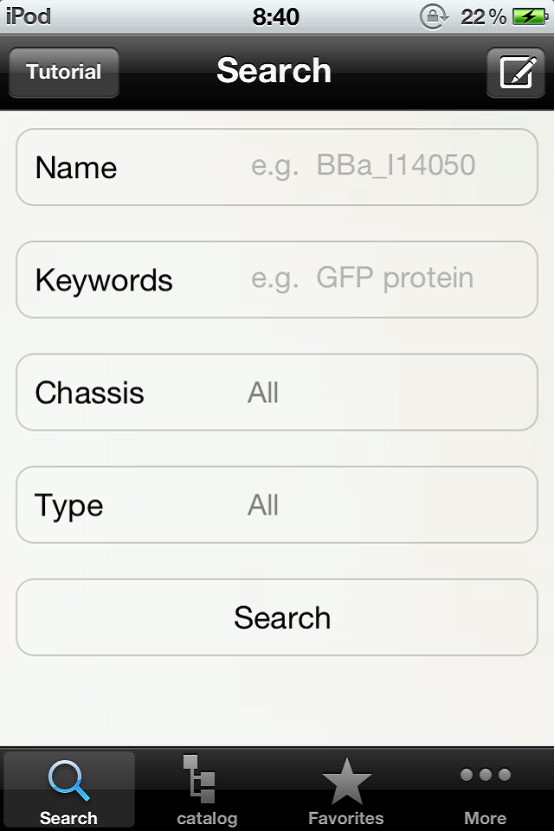
| + | <p> The era of Partsregistry on mobile phone has arrived! With Biosearch on your iPhone, you can now check biobricks and partsregistry in the seminar room; You can design your genetic circuits when you are waiting for a bus! Biosearch is fully interacting with Partsregistry, <a href="http://partsregistry.org">http://partsregistry.org</a>, and has all parts information of Partsregistry database with enhanced user-friendly interface.</p> | ||
| + | <p> Biosearch has a powerful search engine. Users can search parts and devices by type, by category, by keywords, etc. Our online survey shows that Biosearch has major improvement in search result ranking. In addition, our iPhone App has new functions including sharing, rating, adding bookmarks and downloading to local system. These new functions shall promote the communicating and sharing between synthetic biologists. The Biosearch is going to be available on Apple Store and is free to use. </p> | ||
| + | <p> For more information, please click <a href="https://2012.igem.org/Team:SUSTC-Shenzhen-A/Human_practice/Project">here</a>.</p> | ||
| + | <br/> | ||
| + | <p class="title1"> Biodesign</p> | ||
| + | <p><img src="https://static.igem.org/mediawiki/2012/a/a6/Sustc_shenzhen_a_biodesign_overview.png" valign="top" align="right" width="100" style="margin:10px;" /></p> | ||
| + | <p> Our second project, Biodesign, is a graphics editing application specific to synthetic biology for both iPhone and iPad. Biodesign is based on previous iGEM software named Tinkercell and was elaborately improved to adapt to mobile platform. It provides hundreds of biobrick icons as well as lots of fancy functions including dragging graphics, drawing curves and automatically connecting figures with corresponding arrows. With Biosearch, users can draw biochemical reaction schematics anywhere they want and share their drawings with friends. Biodesign will be available online very soon!</p> | ||
| + | <p> For more information, please click <a href="https://2012.igem.org/Team:SUSTC-Shenzhen-A/Biodesign">here</a>.</p> | ||
| + | <p> </p> | ||
| + | <p class="title1"> RFC</p> | ||
| + | <p> We found that the information of many BioBricks is not complete. Thus it is very inconvenient for users to use those information-lacked BioBricks. We want to standardize the minimum information for a qualified BioBrick. If every BioBrick has the same format of information, then it would be very easy to find and distinguish a BioBrick among the sea of BioBricks. In addition, in the website of partsregistry, it is very difficult to find the BioBricks we want due to the useless information in the website. So we conclude that it is essential to create a default template for storing the information of BioBricks. </p> | ||
| + | <p> To see a detailed discussion about this BBF RFC draf, please click <a href="https://2012.igem.org/Team:SUSTC-Shenzhen-A/RFC">here</a>.</p> | ||
| + | <br/> | ||
| + | <h1 class="title"><strong>Sponsors</strong></h1> | ||
| + | <p> </p> | ||
| + | <!--来个table试试--> | ||
| + | <table> | ||
| + | <tr> | ||
| + | <td><a href="http://www.sustc.edu.cn"><img src="https://static.igem.org/mediawiki/2012/2/23/Sustc_sponsor_1.png" width="480" height="80" align="left"/></td> | ||
| + | <td><p>South University of Science and Technology of China Education Fundation</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a href="http://www.crcgas.com"><img src="https://static.igem.org/mediawiki/2012/1/10/Sustc_sponsor_2.jpg" width = "280" height="180"align="left"/></td> | ||
| + | <td><p>Huarun Gas company</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td> | ||
| + | <p> We appreciate helps from many faculties and staffers in SUSTC.</p> | ||
| + | <p> They are Qinshi Zhu, Qin Ye, Wei Han, xuexian wu, Lingjun Wang, Cuiqiong Pang, Jing Zhang, Yun Jiao, Chaoyang Zhu, Yan Zeng and | ||
| + | many others.</p> | ||
| + | <p><strong> Your support is our driving force!</strong> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <h1 class="title"><strong>Acknowledgment</strong></h1> | ||
| + | <p> </p> | ||
| + | <!--来2个table试试--> | ||
| + | <table> | ||
| + | <tr> | ||
| + | <td><a href="https://2012.igem.org/Team:HKUST-Hong_Kong"><img src="https://static.igem.org/mediawiki/2012/1/14/HKUST.png" width="300" height="100" align="left"/></td> | ||
| + | <td><p>We genuinely appreciate iGEM team from Hong Kong University of Science and Technology to have academic communication with us and provide us with precious suggestions and advice.</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a href="https://2012.igem.org/Team:Shenzhen"><img src="https://static.igem.org/mediawiki/2012/7/7e/BGI.png" width="300" height="100"align="left"/></td> | ||
| + | <td><p>We equally genuinely appreciate iGEM team from BGI to have academic communication with us several times and provide us with precious suggestions and advice.Here we express our thanks especially to Kangkang.</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a href="https://2012.igem.org/Team:Tsinghua"><img src="https://static.igem.org/mediawiki/2012/a/ae/Tsinghua.png" width="300" height="100"align="left"/></td> | ||
| + | <td><p>We would also like to give our thanks to iGEM team from Tsinghua University who participate in our survey and give us remarks which truly help us make improvements in our iPhone App.</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a href="https://2012.igem.org/Team:OUC-China"><img src="https://static.igem.org/mediawiki/2012/d/dd/OUC.png" width="300" height="100"align="left"/></td> | ||
| + | <td><p>Thanks to iGEM team from Ocean University of China, we received very good user review about our iPhone App.</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a href="https://2012.igem.org/Team:Tianjin"><img src="https://static.igem.org/mediawiki/2012/b/be/Tianjin.png" width="300" height="100" align="left"/></td> | ||
| + | <td><p>iGEM team from Tianjin University have participated in our survey and send us pieces of constructive advice.Thank you all.</p></td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td><a href="https://2012.igem.org/Team:SUSTC-Shenzhen-B"><img src="https://static.igem.org/mediawiki/2012/1/16/Photo_WIKI2.png" width="300" height="100" align="left"/></td> | ||
| + | <td><p>Here we give our special thanks to team SUSTC-Shenzhen-B. We always grow hand in hand, shoulder by shoulder. We hope both of us can achieve what we deserve. </p></td> | ||
| + | </tr> | ||
| - | |||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| - | |||
| + | <!-- end of header --> | ||
| - | + | </body> | |
| - | + | </html> | |
| - | + | [[file:footbar.jpg|center]] | |
| - | + | ||
Latest revision as of 03:45, 27 October 2012
Project OverviewBiosearch
The era of Partsregistry on mobile phone has arrived! With Biosearch on your iPhone, you can now check biobricks and partsregistry in the seminar room; You can design your genetic circuits when you are waiting for a bus! Biosearch is fully interacting with Partsregistry, http://partsregistry.org, and has all parts information of Partsregistry database with enhanced user-friendly interface. Biosearch has a powerful search engine. Users can search parts and devices by type, by category, by keywords, etc. Our online survey shows that Biosearch has major improvement in search result ranking. In addition, our iPhone App has new functions including sharing, rating, adding bookmarks and downloading to local system. These new functions shall promote the communicating and sharing between synthetic biologists. The Biosearch is going to be available on Apple Store and is free to use. For more information, please click here. Biodesign
Our second project, Biodesign, is a graphics editing application specific to synthetic biology for both iPhone and iPad. Biodesign is based on previous iGEM software named Tinkercell and was elaborately improved to adapt to mobile platform. It provides hundreds of biobrick icons as well as lots of fancy functions including dragging graphics, drawing curves and automatically connecting figures with corresponding arrows. With Biosearch, users can draw biochemical reaction schematics anywhere they want and share their drawings with friends. Biodesign will be available online very soon! For more information, please click here.
RFC We found that the information of many BioBricks is not complete. Thus it is very inconvenient for users to use those information-lacked BioBricks. We want to standardize the minimum information for a qualified BioBrick. If every BioBrick has the same format of information, then it would be very easy to find and distinguish a BioBrick among the sea of BioBricks. In addition, in the website of partsregistry, it is very difficult to find the BioBricks we want due to the useless information in the website. So we conclude that it is essential to create a default template for storing the information of BioBricks. To see a detailed discussion about this BBF RFC draf, please click here. Sponsors
Acknowledgment
|
 "
"