Team:University College London/LabBook/Week13
From 2012.igem.org
YanikaBorg (Talk | contribs) (→Monday 03/09) |
(→Monday 03/09) |
||
| Line 68: | Line 68: | ||
| - | + | {| class = "wikitable" | |
| - | Date Time Colony diameter Halo diameter Absorbance at OD600 | + | !Date !! Time !! Colony diameter !! Halo diameter !! Absorbance at OD600 |
| - | 03.09.2012 12.30pm 0 0 0 | + | |- |
| - | 03.09.2012 16.30pm 0 0 0 | + | |03.09.2012 || 12.30pm || 0 || 0 || 0 |
| - | 03.09.2012 19.30 pm 0 0 0 | + | |- |
| - | 03.09.2012 22.30 pm 0.5mm 1mm 0.002 | + | |03.09.2012 || 16.30pm || 0 || 0 || 0 |
| - | 04.09.2012 01.30 am 1mm 3.5mm 0.182 | + | |- |
| - | 04.09.2012 04.30 am 1.5mm 7mm 0.238 | + | |03.09.2012 || 19.30 pm || 0 || 0 || 0 |
| - | 04.09.2012 07.30 am 2mm 9mm 0.297 | + | |- |
| - | 04.09.2012 10.30am 2.5mm 11mm 0.518 | + | |03.09.2012 || 22.30 pm || 0.5mm || 1mm || 0.002 |
| - | 04.09.2012 12.30pm 3mm 12.5mm 0.701 | + | |- |
| - | 04.09.2012 14.30pm 3.5mm 14mm 0.811 | + | |04.09.2012 || 01.30 am || 1mm || 3.5mm || 0.182 |
| + | |- | ||
| + | |04.09.2012 || 04.30 am || 1.5mm || 7mm || 0.238 | ||
| + | |- | ||
| + | |04.09.2012 || 07.30 am || 2mm || 9mm || 0.297 | ||
| + | |- | ||
| + | |04.09.2012 || 10.30am || 2.5mm || 11mm || 0.518 | ||
| + | |- | ||
| + | |04.09.2012 || 12.30pm || 3mm || 12.5mm || 0.701 | ||
| + | |- | ||
| + | |04.09.2012 || 14.30pm || 3.5mm || 14mm 0.811 | ||
04.09.2012 16.30pm 4mm 15mm 0.906 | 04.09.2012 16.30pm 4mm 15mm 0.906 | ||
Revision as of 03:16, 27 September 2012



Contents |
Monday 03/09
Aim:
To repeat the DNAse/Nuclease Halo experiment in order to check whether results are repeatable. This included collecting data of halo diameters and colony diameters over 28 hour, and to determine how the nuclease production increases overtime.
Methods:
In order to be able to characterise cells, the following protocol was used:
1. Prepare 11-16 plates (10ml DNAse - Agar + 10ul AMP + 10ul 1M IPTG). IPTG induces the lac promoter which in turn activates the transcription of nuclease.
2. Streak cells onto all plates at the same time
3. Incubate at 37°C
4. Apply hydrochloric acid (HCl) to the first plate before putting in the incubator (set time as zero)
5. Take a second reading after four hours, followed by six readings every 3 hours, and a final three readings every two hours.
6. When the reading is taken, observe the following:
a) Diameter of the colony (once the diameter of the colony is measured, pick the colony and put it to grow in LB for nine hours)
b) Diameter of the halo that is achieved once HCl is applied
c) OD from a)
d) Estimate of the depth of the colony on the agar plate
For BL21 cell line that has been modified to contain nuclease:
1. Prepare 11-16 plates (DNAse - Agar + CMP)
2. Streak cells onto all plates at the same time
3. Incubate at 37°C
4. Apply HCl to the first plate before putting in the incubator (set time as zero)
5. Take a second reading after four hours, followed by six readings every 3 hours, and a final three readings
every two hours.
6. When the reading is taken, observe the following:
a. Diameter of the colony (once the diameter of the colony is measured, pick the colony and put it to grow in LB for nine hours)
b. Diameter of the halo that is achieved once HCL is applied
c. OD from a)
d. Estimate of the depth of the colony on the agar plate
Results:
The following table images show the results of the DNase assay:
| Date | Time | Colony diameter | Halo diameter | Absorbance at OD600 | |||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 03.09.2012 | 12.30pm | 0 | 0 | 0 | |||||||||||||||||||||||||||||
| 03.09.2012 | 16.30pm | 0 | 0 | 0 | |||||||||||||||||||||||||||||
| 03.09.2012 | 19.30 pm | 0 | 0 | 0 | |||||||||||||||||||||||||||||
| 03.09.2012 | 22.30 pm | 0.5mm | 1mm | 0.002 | |||||||||||||||||||||||||||||
| 04.09.2012 | 01.30 am | 1mm | 3.5mm | 0.182 | |||||||||||||||||||||||||||||
| 04.09.2012 | 04.30 am | 1.5mm | 7mm | 0.238 | |||||||||||||||||||||||||||||
| 04.09.2012 | 07.30 am | 2mm | 9mm | 0.297 | |||||||||||||||||||||||||||||
| 04.09.2012 | 10.30am | 2.5mm | 11mm | 0.518 | |||||||||||||||||||||||||||||
| 04.09.2012 | 12.30pm | 3mm | 12.5mm | 0.701 | |||||||||||||||||||||||||||||
| 04.09.2012 | 14.30pm | 3.5mm | 14mm 0.811
04.09.2012 16.30pm 4mm 15mm 0.906
 Tuesday 04/09Aim:
Colony Picking
Step 1 – Creating culture media: In a sterile environment, set up X numbers of falcons, each with 5mls of media.
Wednesday 05/09Step 1 - Aim:
MiniPrep Step 1 - Pellet Cells: Pellet 1.5-5ml bacterial culture containing the plasmid by centrifugation g = 6000 Time = 2 min Temperature = (15-25oC) Step 2 - Resuspend Cells: Add 250ul S1 to the pellet and resuspend the cells completely by vortexing or pipetting. Transfer the suspension to a clean 1.5ml microcentrifuge tube. Step 3 - Puncturing Cell Membrane: Add 250ul S2 gently mix by inverting the tube 4-6 times to obtain a clear lysate. Incubate on ice or at room temperature for NOT longer than 5 min. Step 4 - Neutralising S2: Add 400ul Buffer NB and gently mix by inverting the tube 6-10 times, until a white precipitate forms. Step 5 - Centrifuge: g: 14000 Time:10 minutes Temperature: (15-25oC) Step 6 - Centrifuge: Transfer the supernatant into a column assembled in a clean collection tube (provided. Centrifuge: g = 10,000 Time: 1 minute Temperature: (15-25oC) Step 7 - Wash Column: Discard the flow-through and wash the spin column by adding 700ul of Wash Buffer. Centrifuge: g - 10,000 Time - 1 minute Temperature: (15-25oC) Step 8 - Remove residual ethanol: Centrifuge: g - 10,000 Time - 1 minute Temperature: (15-25oC) Step 9 - Elute DNA: Place the column into a clean microcentrifuge tube. Add 50-100ul of Elution Buffer, TE buffer or sterile water directly onto column membrane and stand for 1 min. Centrifuge: g - 10,000 Time - 1 minute Temperature: (15-25oC) Step 10 - Storage: Store DNA at 4oC or -20oC
In order to carry out an analytical digest, the following protocol was used:
Analytical Digest
Step 1 - Thawing cells: Thaw all materials on ice
Step 2 - Adding Ingredient: Add the following ingredients to autoclaved/sterile eppendorf tubes Insert Table Step 3 - Addition of BioBrick: Flick contents gently and centrifuge. Step 4 - Centrifuge: RPM: 14000 Time: 1 minute Temperature: 18oC Step 5 - Digest Program: Place the samples on a thermocycler under the following conditions: RPM: 550 Time: 1 hour Temperature: 37oC Step 6 - Denaturing Enzymes: If you are not running the samples on a gel immediately, denature the restriction enzymes by running the samples on a thermocycler under the following conditions: RPM: 550 Time: 25 minutes Temperature: 65oC Temperature: 18oC
Gel electrophoresis Preparing the Gel Step 1: Within a conical flask, add 3ml 50X TAE, 1.5g Agarose, and 150ml RO water Step 2: Heat for 1 min in microwave. Swirl. Heat again for 30s. If solution is clear stop. Else repeat. Step 3: Cool solution under running cold water. Step 4: Add 20ul ethidium bromide (normal concentration of EB solution is 500ug/ul) Step 5: Pour into a sealed casting tray in a slow steady stream, ensuring there are no bubbles Running a gel Step 6: Add 1 part loading buffer to five parts of loading sample Step 7: Position the gel in the tank and add TAE buffer, enough to cover the gel by several mm Step 8: Add 5ul of DNA ladder to lane 1 Step 9: Add samples to the remaining wells Step 10: Run at 100 volts for 1hour and 15 minutes Imaging the Gel Step 11: Place gel in GelDoc 2000 chamber Step 12: Turn GelDoc 2000 chamber on Step 13: From computer: Quantity One > Scanner > Gel_Doc_Xr>Manuqal Acquire Step 14: Alter the exposure/settings to give a clear image. TAE - Tris-acetate-EDTA EDTA - ethylenediamine tetraacetic acid
Conclusion:
Step 3 - Aim:
X+P cut to E+P cut Plasmid All. X+S Plasmid backbone religated control and no ligase control. E+P cut ire to E+P cut plasmid backbone.
Thursday 06/09Aim:

Wednesday 05/09Aim: To characterise the two strains of laccase obtained from the laccase ligation.
Laccase characterisation In order to characterise laccase, the following steps are carried out:
 Thursday 06/09Aim:
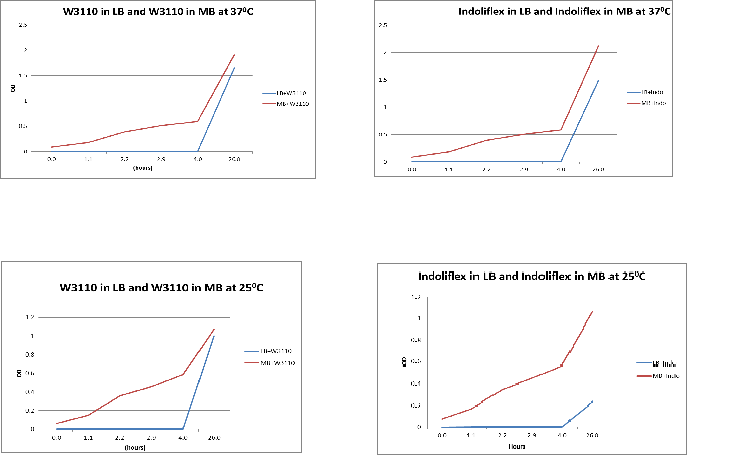
Growth rate characterisation In order to compare growth rates of W3110 and indolifex in LB and MB media, the following steps were carried out:
Results:
Conclusion:
|
 "
"