Team:TU Darmstadt/Labjournal/Degradation
From 2012.igem.org
(→Thursday, 29.05.12) |
(→Thursday, 26.04.12) |
||
| Line 353: | Line 353: | ||
* [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] (Agarose gele elektrophoresis) for qualitiy control | * [https://2012.igem.org/Team:TU_Darmstadt/Protocols/Agarose_gel_electrophoresis Agarose gel electrophoresis] (Agarose gele elektrophoresis) for qualitiy control | ||
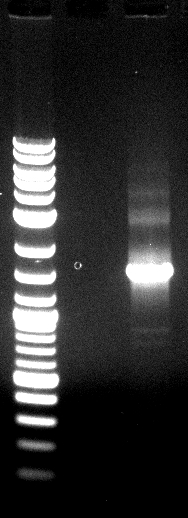
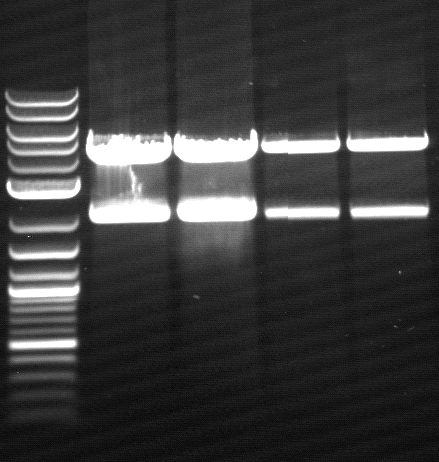
[[File: 120426_PCR_1-3_siehe_Laborbuch_26.4-1.png]] | [[File: 120426_PCR_1-3_siehe_Laborbuch_26.4-1.png]] | ||
| - | * 2-12= | + | * 2-12= phoA |
==== Friday, 27.04.12 ==== | ==== Friday, 27.04.12 ==== | ||
Revision as of 20:56, 26 September 2012
Degradation
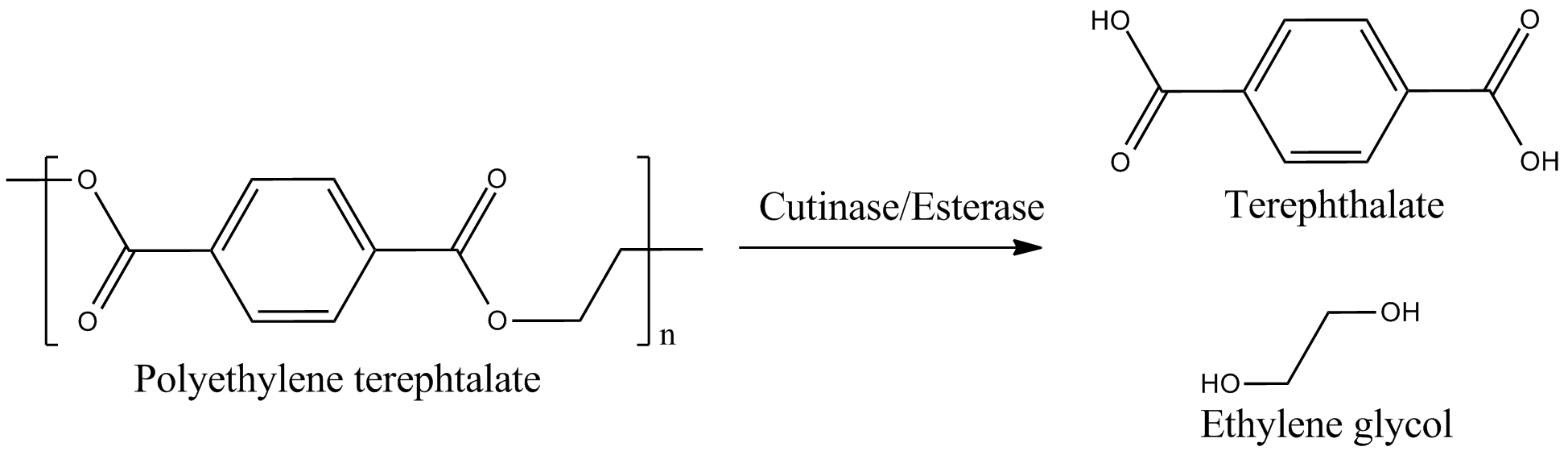
This page features the work carried out by the degradation team. The main objectives were the production of a BioBrick containing Fusarium solani cutinase or Est13 esterase two enzymes potentially enabling E.coli of PET degradation and over-expression stems for activity screening.
The degradation group consists of six undergraduates and two PhD student advisors. Our objective is the expression of a fusion protein on the surface of E. coli to enable a microbial polyethylenterephtalate (PET) degradation.
We identified three potential PET degradation enzymes from literature. Two of them are cutinases HiC (Humicola insolens cutinase) and FsC (Fusarium solani cutinase), the other namely pNB-Est13 beeing an esterase. After a short examination we dropped the HiC due to a temperature optimum of 80+°C. Shortly after the FsC was dropped as well, due to its toxicity for E.coli.In order to maximise the activity we decided to anchor and display the cutinase/esterase directly on the surface of the producing bacterial cell. Surface-exposed enzymes are directly accessible to the respective substrates which no longer have to traverse the cellular membrane barriers. Furthermore, the enzyme reaction occurs in a chemically more defined environment as compared to the interior of a microbial cell. We use the outer membrane proteins of Pseudomonas aeruginosa (EstA) as a membrane anchor and the signaling sequence of PhoA translocators to display the enzyme on the outer surface of E. coli cells. In addition the fusion protein contains a his-tag and a myc-tag for detection via flow cytometry after antibody staining or Western blot.The degradation group consists of six undergraduates and two PhD student advisors. Our objective is the expression of a fusion protein on the surface of E. coli to enable a microbial polyethylenterephtalate (PET) degradation.
At first the signal sequence (PhoA), the catalytic domain (FsC/Est13) and EstA were assembled inline from their respective vectors. This is due to the fact, that by combining multiple parts in the standardized BioBrick vectors, scars with stop codons are generated that would effectively prevent the fusionproteins expression. In consideration of the DNA constructs length we pursued two PCR based synthesis strategies. One being the SKV (standard cloning procedure) the other being the SOE (standard overlap procedure). The first using primers and restriction sites for assembly the latter using overlapping primers. During both assembly procedures restriction sites of PstI, EcorI, SpeI or XbaI were eliminated from the coding sequence by mutagene PCR. In the end we completely changed our assembly strategy, using synthesis products and BsaI sites to put our parts together. After completion the fusionprotein and its subunits were transfered to the BioBrick standard and sent to the registry. For further characterisation the enzymes were overexpressed in E. coli strains Top10, DH5α, Mg1655, screened on tributyrin agar and detected via flow cytometry after performing an antibody staining. The material science group went even further and tried to examine them using AFM.
Contents
|
SOE PCR
SOE PCR stands for Splicing by Overlapp Extension PCR. It is a standard overlapp extension procedure, enabling the assembly of genes wihtout performing any cloning, digesting or ligation inbetween. All you need to do ist running PCRs with specific primers. If gene A is the upstream part and gene B has to be assembled downstream of gene A, primer lo of gene A should have an overlapp of around 20 nucleotides complementary to the first 20 nucleotides of gene B. Primer up of gene B should haven a complementary overlapp of 20 nucleotides to the end of gene A.
We stopped using SOE PCR after a couple of weeks. It is a good method to perform mutationial PCR but to assemble genes of the length of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808032 BBa_K808032] it is not the best. Due to bad yields of gel extraction after the first assembly steps we stopped SOE PCR and went on with [1]. But eventually we got our chimeric genes synthesized by [http://de-de.invitrogen.com/site/de/de/home/Products-and-Services/Applications/Cloning/gene-synthesis/GeneArt-Gene-Synthesis.html GeneArt].
Week 1 / CW 17
Tuesday, 24.04.12
- Production of electrocompetent cells [http://ecoliwiki.net/colipedia/index.php/DH5_alpha DH5alpha] and [http://ecoliwiki.net/colipedia/index.php/Category:Strain:BL21 BL21]
- Pouring of LB-Agar plates with ampecilin resistance (AMP)
- setting of DYT media
- Electroporation of BL21 with the following plasmids
- pEST100, carrying a CMP-resistance, the genes of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808028 phoA], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808025 FsC] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808027 EstA]
- pET26b(+), carrying a Kan-resistance and the gene of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808026 pNB-Est13]
- pET16b, carrying an Amp-resistance , and is needed for our SKV
- transformed Bl21 cells are incubated over night at 37°C in DYT-media and crossed out on LB-Agar plates
Wednesday, 25.04.12
- Miniprep of the 3 overnigth [http://ecoliwiki.net/colipedia/index.php/Category:Strain:BL21 Bl21] cultures and concentration meassurement via Nanodrop
Thursday, 26.04.12
Annotation: Every PCR is done in 4 assays à 50 µL, TA = 60°C, ta = 35s, tE = 25 s
- PCR on pEST100
- PCR on pEST100
- gene of interest: Promo-LacO-RBS-Phoa for SOE PCR with pNB-Esterase13
- primers: SOE A up & SOE a1 lo
- PCR on pEST100]
- gene of interest: Promo-LacO-RBS-Phoa for SOE PCR with FsC
- primers: SOE A up & SOE a2 lo
- PCR on pEST100
- gene of interest: FsC for SOE PCR with Promo-LacO-RBS-Phoa and EstA part1
- primers: SOE b2 up & SOE b2 lo
- PCR on pET26b(+)
- gene of interest: pNB-Est13 part1 for SOE PCR with Promo-LacO-RBS-Phoa and pNB-Est13 part2
- primers: SOE b1 up & SOE Est13 mut lo
- PCR on pET26b(+)
- gene of interest: pNB-Est13 part2 for SOE PCR with pNB-Est13 part1 and EstA part1
- primers: SOE Est13 mut up & SOE b1 lo
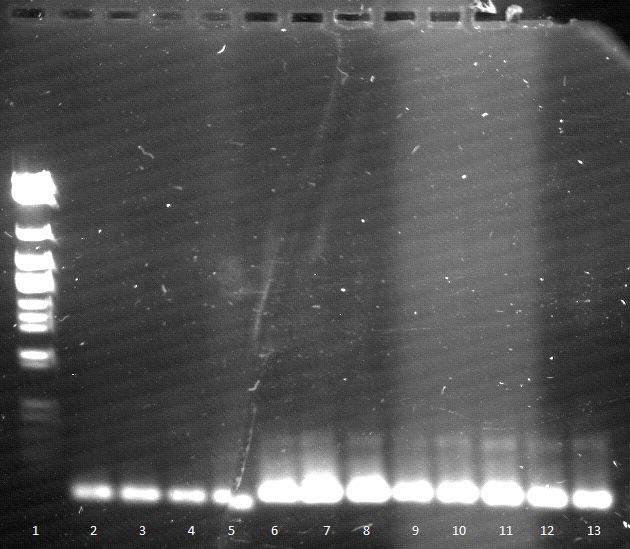
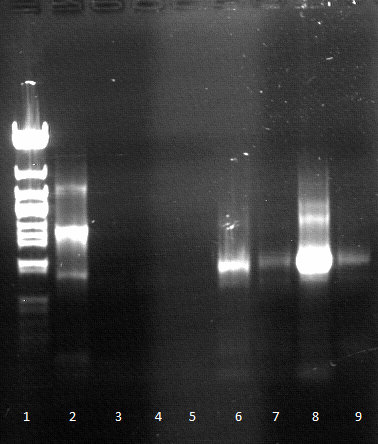
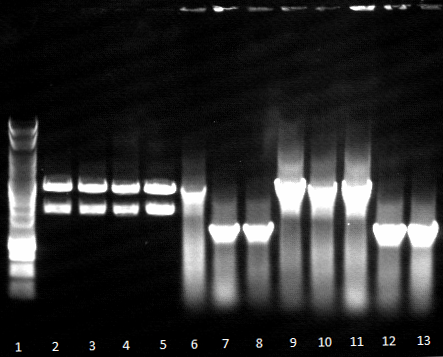
- Agarose gel electrophoresis for qualitiy control
- 2-5 1.PCR, 6-9 2.PCR, 10-13 3.PCR
- 2-5 4.PCR, 6-9 5.PCR, 710-13. 6.PCR
Friday, 27.04.12
- clean up of 1. PCR on pEST100 from Thursday, 26.04.12 via Ammonium acetate - Ehtanol DNA precipitation, solved in 54 µl ddH2O
- restriction of cleaned up 1. PCR and pET16b with XbaI and NdeI over weekend at 37°C
- Annotation: From now on SKV will be protocolled in SKV wiki.
Week 2 / CW 18
Monday, 30.04.12
- SOE PCR
- pNB-Est13 part1 & pNB-Est13 part2, primers: SOE b1 up & SOE b1 lo
- Promo-LacO-RBS-Phoa & FsC, primers: SOE A up & SOE b2 lo
- Annotation: Every PCR is done in 4 assays à 50 µL, TA = 60°C, ta = 35s, tE1 = 20s, tE1 = 35s
- Agarose gel electrophoresis of SOE PCR
- SOE PCR of Promo-LacO-RBS-Phoa-FsC worked, pNB-Est13 did not due to missing clean up via Agarose gel electrophoresis, precipitation is insufficient, we do it again
- PCRs
- PCR on pET26b(+)
- gene of interest: pNB-Est13 part1 for SOE PCR with Promo-LacO-RBS-Phoa and pNB-Est13 part2
- primers: SOE b1 up & SOE Est13 mut lo
- PCR on pET26b(+)
- gene of interest: pNB-Est13 part2 for SOE PCR with pNB-Est13 part1 and EstA part1
- primers: SOE Est13 mut up & SOE b1 lo
- Agarose gel electrophoresis
- 1. PCR worked well 2. PCR did not
Wednesday, 02.05.12
- PCRs
- PCR on pEST100
- gene of interest: Promo-LacO-RBS-Phoa for SOE PCR with pNB-Est13
- primers: SOE A up & SOE a1 lo
- PCR on pET26b(+)
- gene of interest: pNB-Est13 part1 for SOE PCR with Promo-LacO-RBS-Phoa and pNB-Est13 part2
- primers: SOE b1 up & SOE Est13 mut lo
- PCR on pET26b(+)
- gene of interest: pNB-Est13 part2 for SOE PCR with pNB-Est13 part1 and EstA part1
- primers: SOE Est13 mut up & SOE b1 lo
- TA = 57°C, ta = 35s, tE = 25 s
- every PCR is performed in 3 batches à 50 µL
- Agarose gel electrophoresis
- Gel extraction
- c(1.PCR)=6 ng/µL
- c(2.PCR)=12 ng/µL
- c(3.PCR)=13 ng/µL
- SOE PCR of 2.PCR and 3.PCR in order to form pNB-Est13 for SOE PCR with Promo-LacO-RBS-Phoa and EstA
- TA1 = 60°C, ta1 = 25 s, tE1 = 25 s
- TA = 60°C, ta2 = 25 s, tE2 = 35 s
Tuesday, 03.05.12
- preparative Agarose gel electrophoresis of SOE PCR from yesterday
- Gel extraction
- c(pNB-Est13) 7 ng/µL
Friday, 04.05.12
- PCRs
- PCR on pEST100
- gene of interest: Promo-LacO-RBS-Phoa for SOE PCR with pNB-Est13
- primers: SOE A up & SOE a1 lo
- PCR on pEST100
- gene of interest: EstA part1 for SOE PCR with Promo-LacO-RBS-Phoa and pNB-Est13
- primers: SOE c1 up & SOE EstA mut PstI out lo
- PCR on pEST100
- gene of interest: EstA part1 for SOE PCR with Promo-LacO-RBS-Phoa and FsC
- primers: SOE c2 up & SOE EstA mut PstI out lo
- PCR on pEST100
- gene of interest: EstA part2 for SOE PCR with EstA part1
- primers: SOE EstA mut PstI out up & SOE D lo
- annotation: TA = 60°C, ta = 25 s, tE = 35 s
- every PCR is performed in 3 batches à 50 µL
- preparative Agarose gel electrophoresis
Week 3 / CW 19
Tuesday 08.05.12
- gene of interest: Promo-LacO-RBS-Phoa-pNBEst13 for SOE PCR with EstA
- primers: SOE A up & SOE b1 lo
- template: pNB-Est13 from last friday & Promo-LacO-RBS-Phoa from Wednesday, 02.05.12
- annotation: TA1 = 60°C, ta1 = 25 s, tE1 = 35 s
- TA2 = 60°C, ta2 = 25 s, tE2 = 1 min
- did not work
Wednesday, 09.05.12
- PCR on pET26b(+)
- gene of interest: pNB-Est13 part1 for SOE PCR with Promo-LacO-RBS-Phoa and pNB-Est13 part2
- primers: SOE b1 up & SOE Est13 mut lo
- PCR on pET26b(+)
- gene of interest: pNB-Est13 part2 for SOE PCR with pNB-Est13 part1 and EstA part1
- primers: SOE Est13 mut up & SOE b1 lo
- PCR on pEST100
- gene of interest: EstA part1 for SOE PCR with Promo-LacO-RBS-Phoa and pNB-Est13
- primers: SOE c1 up & SOE EstA mut PstI out lo
- PCR on pEST100
- gene of interest: EstA part1 for SOE PCR with Promo-LacO-RBS-Phoa and FsC
- primers: SOE c2 up & SOE EstA mut PstI out lo
- PCR on pEST100
- gene of interest: EstA part2 for SOE PCR with EstA part1
- primers: SOE EstA mut PstI out up & SOE D lo
- annotation: TA = 60°C, ta = 25 s, tE = 35 s
- every PCR is performed in 2 batches à 50 µL
Friday, 09.05.12
- qualitative Agarose gel electrophoresis
Week 4 / CW 20
Monday, 14.05.12
- Gel extraction of remaining PCRs from Wednesday, 09.05.12
- c(pNB-Est13 part1)=24 ng/µL
- c(pNB-Est13 part2)=26 ng/µL
- c(EstA part2)=13 ng/µL
- new SOE EstA mut PstI out lo is orderd from Sigma-Aldrich
- SOE PCR
- gene of interest: pNB-Est13 for SOE PCR with EstA and Promo-LacO-RBS-Phoa
- primers: SOE b1 up & SOE b1 lo
- template: pNB-Est13 part1 from last friday & pNB-Est13 part2
- annotation: TA1 = 52°C, ta1 = 25 s, tE1 = 35 s
- TA2 = 57°C, ta2 = 25 s, tE2 = 1.15 min
- Agarose gel electrophoresis
Tuesday, 15.05.12
- Gel extraction of SOE PCR from yesterday
- c(pNB-Est13)=24 ng/µL
- SOE PCR
- SOE PCR
- gene of interest: Promo-LacO-RBS-Phoa-pNBEst13 for SOE PCR with EstA
- primers: SOE A up & SOE b1 lo
- template: pNB-Est13 from yesterday & Promo-LacO-RBS-Phoa from Wednesday, 02.05.12
- SOE PCR
- gene of interest: Promo-LacO-RBS-Phoa-FsC for SOE PCR with EstA
- primers: SOE A up & SOE b2 lo
- template: FsC from Thursday, 26.04.12 & Promo-LacO-RBS-Phoa from Wednesday, 02.05.12
- annotation: TA1 = 52°C, ta1 = 25 s, tE1 = 35 s
- TA2 = 57°C, ta2 = 25 s, tE2 = 1.15 min
- each PCR is performed in 2 batches à 50 µL
Wednesday, 16.05.12
- SOE PCR of Promo-LacO-RBS-Phoa-pNBEst13 did not work but Promo-LacO-RBS-Phoa-FsC worked
- Gel extraction
- c(Promo-LacO-RBS-Phoa-FsC)=10 ng/µL
Week 5 / Kw 21
Monday, 23.05.12
- PCR on pEST100
- gene of interest: EstA part1 for SOE PCR with pNB-Est13 and EstA part2
- primers: SOE c1 up & SOE EstA mut PstI out lo
- PCR on pEST100
- gene of interest: EstA part1 for SOE PCR with FsC and EstA part2
- primers: SOE c2 up & SOE EstA mut PstI out lo
Tuesday, 22.05.12
- qualitative Agarose gel electrophoresis
- Gel extraction
- c(EstA part1 for SOE PCR with FsC)=4 ng/µL
- c(EstA part1 for SOE PCR with pNB-Est13)=7 ng/µL
Wednesday, 23.05.12
- SOE PCR
- gene of interest: EstA for SOE PCR with FsC
- primers: SOE c2 up & SOE D lo
- template: EstA part1 from yesterday & EstA part2 from Monday, 14.05.12
- annotation: TA1 = 52°C, ta1 = 25 s, tE1 =45 s
- TA2 = 65°C, ta2 = 25 s, tE2 = 90 s
- qualitative Agarose gel electrophoresis
- Gel extraction
- c(EstA for SOE PCR with pNB-Est13)=42 ng/µL
- c(EstA for SOE PCR with FsC)=23 ng/µL
Thursday, 24.05.12
- PCR on EstA for SOE PCR with pNB-Est13 from yesterday
- gene of interest: EstA for SOE PCR with pNB-Est13
- primers: SOE c1 up & SOE D lo
- PCR on EstA for SOE PCR with FsC
- gene of interest: EstA for SOE PCR with FsC
- primers: SOE c2 up & SOE D lo
- PCR on pEST100
- gene of interest: Promo-LacO-RBS-Phoa for SOE PCR with pNB-Est13
- primers: SOE A up & SOE a1 lo
- PCR on pEST100
- gene of interest: FsC for SOE PCR with Promo-LacO-RBS-Phoa and EstA part1
- primers: SOE b2 up & SOE b2 lo
- annotation: TA = 60°C, ta = 25 s, tE =45 s
- each PCR is performed in 3 batches à 50 µL
- qualitative Agarose gel electrophoresis
- both EstAs worked
- Promo-LacO-RBS-Phoa for SOE PCR with pNB-Est13 worked
- Gel extraction
- c(EstA for SOE PCR with pNB-Est13)=39 ng/µL
- c(EstA for SOE PCR with FsC)=20 ng/µL
- c(Promo-LacO-RBS-Phoa for SOE PCR with pNB-Est13)=13 ng/µL
Week 6 / Kw 22
Tuesday, 29.05.12
- SOE PCR
- gene of interest: Promo-LacO-RBS-Phoa-Fsc-EstA
- primers: SOE A up & SOE D lo
- template: EstA from Thursday, 24.05.12 & Promo-LacO-RBS-Phoa-Fsc from Wednesday, 16.05.12
- SOE PCR
- gene of interest: Promo-LacO-RBS-Phoa-pNBEst13-EstA
- primers: SOE A up & SOE D lo
- template: EstA from Thursday, 24.05.12 & Promo-LacO-RBS-Phoa from Wednesday, 02.05.12 & pNB-Est13 from Monday, 14.05.12
- SOE PCR
- gene of interest: Promo-LacO-RBS-Phoa-pNBEst13 for SOE PCR with EstA
- primers: SOE A up & SOE b1 lo
- template: Promo-LacO-RBS-Phoa from Wednesday, 02.05.12 & pNB-Est13 from Monday, 14.05.12
- annotation: each PCR is performed in 3 batches à 50 µL
- TA1 = 52°C, ta1 = 25 s, tE1 =45 s
- TA2 = 65°C, ta2 = 25 s, tE2 = 90 s
- Agarose gel electrophoresis
- Promo-LacO-RBS-Phoa-Fsc-EstA worked
- Promo-LacO-RBS-Phoa-pNBEst13 worked
Thursday, 29.05.12
- Gel extraction
- c(Promo-LacO-RBS-Phoa-Fsc-EstA)= 8 ng/µL
- c(Promo-LacO-RBS-Phoa-pNBEst13)= 13 ng/µL
Due to better results in SKV we stopped our tries in SOE PCR assembly of our chimeric protein [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808030 BBa_K808030].
SKV
Week 1 / CW 17
Tuesday, 24.04.12
- Production of electrocompetent cells DH5alpha and [http://ecoliwiki.net/colipedia/index.php/Escherichia_coli BL21]
- Pouring of LB-Agar plates with ampecilin resistance (AMP)
- setting of DYT media
- Electroporation of BL21 with the following plasmids
- pEST100, carrying a CM-resistance, the genes of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808028 phoA], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808025 FsC] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808027 EstA]
- pET26b(+), carrying a Kan-resistance and the gene of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808026 pNB-Est13]
- pET16b, carrying an Amp-resistance , and is needed for our SKV
Wednesday, 25.04.12
- Miniprep of the 3 overnigth [http://ecoliwiki.net/colipedia/index.php/Escherichia_coli BL21] cultures and concentration meassurement via Nanodrop
Thursday, 26.04.12
Annotation: Every PCR is done in 4 assays à 50 µL, TA = 60°C, ta = 35s, tE = 25 s
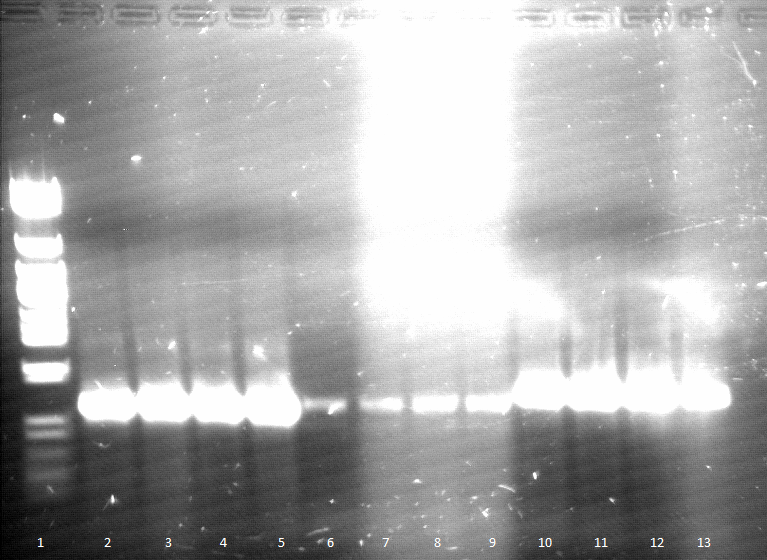
- PCR on pEST100
- Agarose gel electrophoresis (Agarose gele elektrophoresis) for qualitiy control
- 2-12= phoA
Friday, 27.04.12
- clean up of 1. PCR on pEST100 from Thursday, 26.04.12 via Ammonium acetate - Ehtanol DNA precipitation, solved in 54 µl ddH2O
- restriction of cleaned up 1. PCR and pET16b with XbaI and NdeI over weekend at 37°C
Week 2 / CW 18
Monday, 30.04.12
- Agarose gel electrophoresis of DNA digestion from Friday, 27.04.12
- Only one single band on Agarose gel electrophoresis
- For saveness two single digests are performed at 20 µL batches. One with XbaI and the other with NdeI. Incubation for 2 hours.
- Agarose gel electrophoresis of single digests
- Enzymes cut once each, so digest should have worked.
- Ligation of PhoA in cut (XbaI / NdeI) pET16b
- Annotation: 30 µL batch, 1:5 ratio, Ligation at 4°C for 2 days till Wednesday, 02.05.12
Week 4/CW 20
Monday, 07.05.
- Electroporation of DH5alpha with 5 µl of the ligation which was performed 2 days before.
- Restriction digest of the remaining PCR product PhoA with XbaI/NdeI for 1,5h at 37°C Thursday, 26.04.
- PhoA is cleaned up by Agarose gel electrophoresis
- concentration:7,36 ng/µl,10,6 ng/µl
- Ligation of PhoA x pET16b for 2h, at the rate of (Vector/Insert) 1:5, 1:3
Annotation: If it does not say anything else, Ligation is always done in 20µl batches.
Tuesday, 08.05.
- Transformation of DH5alpha with PhoA x pET16b was not successfull, so it was performed again Monday, 07.05.
- DNA digestion of pET16b
- 100 µl are cut with XbaI/NdeI, incubaton-time: 1,5h at 37°C
- product is cleaned up by Ammonium acetate - Ehtanol DNA precipitation
- concentration: 85 ng/µl
Wednesday, 09.05.
- Colony-PCR
- gene of interest: PhoA
- primer:SKV a1 up XbaI, SKV a1 lo NdeI
- PhoA was not inserted, but 2 colonies were picked and inoculated in 5 ml LB Amp medium to look at it again due to a test-restriction.
Thursday, 10.05.
- Miniprep of PhoA x pET16b
- Restriction digest of the Miniprep: (PhoA x pET16b)
- enzymes: XbaI/NdeI, incubation: 1,5 h, 37°C
- digest shows that PhoA was not inserted
- Ligation is performed again Monday, 07.05.
- Amplification of the genes FSC, Est13 and EstA via PCR
- templates:
- FSC: pEST100
- Est13: pET26b
- EstA: product of SOE-PCR
- primer:
- SKV b1 up NdeI, SKV b1 lo NcoI
- SKV b2 up NdeI, SKV b2 lo NCOI
- SKV c1 up NcoI, SKV c1 lo EcoRI
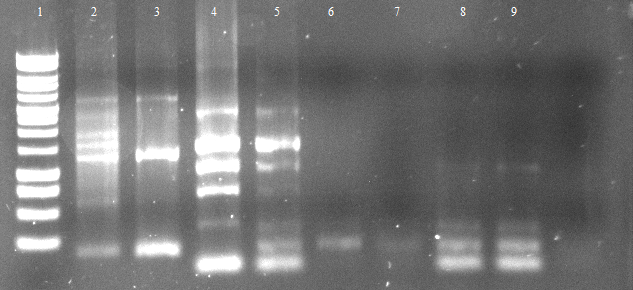
Friday, 11.05.
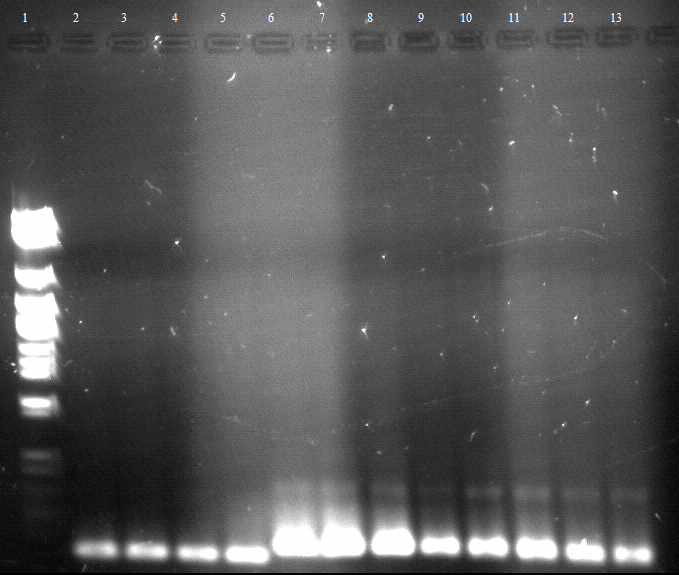
- an analytic, 1% agarose-gel proves that the amplification of FSC, Est13 and EstA was successfull.
- But to amplify Est13 it was used the product of the SOE PCR which had not been cleaned up, so the large Fragments of the SOE-primers might have disturbed the amplification. The PCR of Est13 had to be performed again (Wednesday, 16.05.)
- 2,3,5,6= Est13
- 7,8,9,10= EstA
- 11,12= FSC
- DNA digestion of pET16b to produce template for a new ligation of PhoA
- concentration: 76,3 ng/µl
- enzymes: XbaI, NdeI
- incubation: for 2 days, 37°C
- Ligation of PhoA x pET16b cut with XbaI and NdeI is carried out using different rates of Vector and insert:
- ratio vector/insert= 3:1, 1:1, 1:3, 1:5, 1:7, 1:10
- PhoA= 10,6 ng/µl Monday,07.05.
- incubation: 2 days, 4°C
Week5/CW21
Monday, 14.05.
- the Restriction digest of pET16b cut with XbaI and NdeI is isolated by Ammonium acetate - Ehtanol DNA precipitation
- Ligation of (phoA x pET16b) is used to transform DH5alpha via Electroporation
Tuesday, 15.05.
- Colony-PCR
- primer:
- SKV a1 up XbaI,
- SKV a1 lo NdeI
- PhoA is inserted, so 10 clones (2-9,11,12) are picked to be grown in 5ml DYT over night.
Wednesday, 16.05.
- Miniprep of 10 clones, the plasmid-DNA is isolated out of 3ml culture per clone.
- Restriction digest with NdeI and NcoI of the plasmids (phoA x pET16b) 1-4, so FSC and Est13 can be added to the construct.
- FSC is cut with NdeI and NcoI
- digests are performed in 20µl total volume at 37°C for 1,5h and cleaned up by Agarose gel electrophoresis
- PCR of EST13
- template: product of SOE-PCR, cleaned up by the Promega-Kit
- primer: SKV b1 up NdeI, SKV b1 lo NcoI
- an analytical Agarose gel electrophoresis is performed
- 7-10 = Est 13
Friday, 18.05.
- Ligation of FSC x (phoA x pET16b)
- ratio vector/insert: 3:1, 1:1, 1:3
- concentration:
- (phoA x pET16b)= 3,72 ng/µl
- FSC= 12,22 ng/µl
Week 6/CW 22
Monday, 21.05.
- Elektroporation of DH5alpha with the Ligation of (FSC) x (phoA x pET16b)
- Restriction digest of (phoA x pET16b)(150µl,160µl) for the insertion of Est13/FSC
- enzymes: NdeI, NcoI
- incubation: 1,5h, 37°C
- cleaned up via Agarose gel electrophoresis
- the DNA fragment at 600 bp proves that the vector has been cut with NdeI and NcoI.
- Restriction digest of Est13
- cleaned up via Agarose gel electrophoresis
- c(Est13)= 7,7 ng/µl
Tuesday, 22.05.
- Ligation of Est13 and (phoA x pET16b)
- both components have been cut with NdeI and NcoI Monday, 21.05.
- ratio vector/insert: 1:10, 1:5, 1:3
- incubation: 2h, 25°C
- Electroporation of dH5alpha using the Ligation of Est13 x (phoA x pET16b), 5 µl per 100µl cells
Wednesday, 23.05.
- Colony-PCR
- aim is to check the insertion of Est13
- Primer:SKV b1 up NdeI, SKV b1 lo NcoI
- 8 colonies have been picked and the Agarose gel electrophoresis proves, that Est 13 has not been inserted into (phoA x pET16b
- Ligation of FSC and Est13 in (phoA x pET16b)
- ratio vector/insert: 1:3, 1:5
- c(templates):Monday, 21.05.
- Elektroporation of DH5alpha with Est13 in (phoA x pET16b)
Thursday, 24.05.
- Colony-PCR proves, that Est13 has been inserted into (phoA x pET16b)
- plates are covered with colonies, 8 colonies are chosen for colony-PCR
- colony No.5 is chosen to be inoculated in 5µ DYT/Amp medium
- PCR is performed again to amplify FSC and Est13
- products are cleaned up by Agarose gel electrophoresis
Friday, 25.05.
- Restriction digest of FSC and Est13 Thursday, 24.05.
- enzymes: NdeI, NcoI
- incubation: 2days, 37°C
Week 7/CW 23
Tuesday, 29.05.
- Restriction digest of FSC and Est13 is cleaned up via Agarose gel electrophoresis
- Miniprep of Est13 x (phoA x pET16b) via Promega-Kit
- Ligation of FSC x (phoA x pET16b)
- ratio vector/insert: 1:3, 1:5, 1:10
- incubation: 2h, 25°C
- Electroporation of DH5alpha with 5µl of the Ligaton: FSC x (phoA x pET16b)
- PCR to amplify EstA, (Thursday, 10.05.)
- 4 assays à 50 µL, TA = 66°C, ta = 35s, tE = 90s
- cleaned up via Kit
- An analytical electrophoresis is performed
Wednesday, 30.05.
- Restriction digest of EstA (product of PCR, 80 µl) and Plasmid: (Est13 x phoA x pET16b)
- enzymes: EcoRI, NcoI
- incubation: 1,5h, 37°C
- The Agarose gel electrophoresis shows, that two DNA fragments are cut out of the vector (Est13 x phoA x pET16b). This might be a hint that Est13 is not inserted.
- 1-3 = (Est13 x phoA x pET16b)
- 4-5 = EstA
- c(EstA) = 6,7 ng/µl
- c(Est13 x phoA x pET16b)= 23 ng/µl
- Electroporation of (FSC x phoA x pET16b) worked well, plates are covered with colonies which are picked to be grown in 5ml DYT/Amp medium over night.
Thursday, 31.05.
- Miniprep of the amplified (FSC x phoA x pET16b)
- Restriction digest to ligate EstA
- enzymes:NcoI, EcoRI
- incubation: 1,5h, 37°C
- to be sure that the plasmid (Est13 x phoA x pET16b) really carries Est13 and PhoA, an analytic PCR is performed. Also pEt16b is used as template with different primers.
- templates:
- pET16b Wednesday, 25.04.
- (phoA]] x pET16b) Wednesday, 16.05.
- (Est13 x phoA x pET16b) Tuesday,29.05.
- Primers, used on each template:
- SKV b1 up NdeI, SKV b1 lo NcoI
- SKV a1 up XbaI, SKV a1 lo NdeI
- conditions:
- TA = 66°C, ta = 35s, tE = 90s
- the Agarose gel electrophoresis shows, that there is no binding-specifity concerning the genes of interest and the primers, but primers have been checked on mistakes.
- to solve the problem, the Ligation of FSC and Est13 shall be repeated from the beginning.
- Additionally,the decision was to go on with the Ligation of EstA into the Plasmids (Est13 x phoA x pET16b) and (FSC x phoA x pET16b), in case that the PCR was proceeded under wrong conditions.
Friday, 01.06.
- Restriction digest of (phoA x pET16b)
- enzymes: NcoI, NdeI
- incubation: 1,5 h, 37°C
- Ligation of Est13 and FSC x (PhoA x pET16b)
- ratio vector/insert: 1:3, 1:5
- c([[Est13)= 3 ng/µl
- c(FSC)= 11 ng/µl
- c(phoA x pET16b)= 29 ng/µl
- incubation: over night, 25°C
- Restriction digest of (FSC x (PhoA x pET16b)
- enzymes: NcoI, EcoRI
- incubation: 1,5h, 37°C
Week 8/CW 24
Monday, 04.06.
- restriction of (FSC x (phoA x pET16b) and (phoA x pET16b) is cleaned up by Agarose gel electrophoresis
- c(FSCx (phoA x pET16b) = 14 ng/µl
- Electroporation of DH5alpha with Ligation: (FSC x phoA x pET16b), (EST13 x phoA x pET16b) Friday, 01.06
Tuesday, 05.06.
- Colony-PCR on EST13 & FSC is negative.
Wednesday, 06.06.
- Electroporation of DH5alpha with Ligation-assays from 01.06.
- Ligation of EstA Wednesday,30.05 and the Plasmids (Est13 x phoA x pET16b),(FSC x phoA x pET16b) from Monday,04.06.
- ratio vector/insert: 1:3,1:5
- incubation: 2h, 25°C
- Electroporation of DH5alpha with (EstA x Est13 x phoA x pET16b) and (EstA x FSC x phoA x pET16b)
Friday, 08.06.
- No cells are grown on plates, so the plates are incubated for another two days at RT.
Week8/ CW24
Monday, 11.06.
- all plates are covered with cells, so a colony-PCR is performed on either Est13,FSC or EstA
- Primer:
- SKV b1 up NdeI, SKV b1 lo NcoI
- SKV b2 up NdeI, SKV b2 lo NCOI
- SKV c1 up NcoI, SKV c1 lo EcoRI
- Every PCR is done in 8 assays à 50 µL, TA = 57°C, ta = 35s, tE = 90 s
Tuesday, 12.06.
- Agarose gel electrophoresis shows, that EstA has been inserted in both Plasmids, (Est13 x phoA x pET16b) and (FSC x phoA x pET16b)
- colonies No.3,4 (EstA x FSC x phoA x pET16b) and No.15,16 (EstA x Est13 x phoA x pET16b) seem to be positive on EstA and are picked to be grown in 5ml DYT/Amp medium.
- Colony-PCR on Est13 and FSC shows a lot of unspecific bands, so cloning of the genes from the beginning has to be dropped.
Wednesday, 13.06.
- Miniprep of the clones No. 3,4,15 and 16. Tuesday, 12.06.
- c(EstA x FSC x PhoA x pET16b)=
- No.3=65,17 ng/µl
- No.4=63,8 ng/µl
- c(EstA x Est13 x PhoA x pET16b)=
- No.15=66,3 ng/µl
- No.16=59,7 ng/µl
- for gene-expression, it is performed a elektroporation of [http://ecoliwiki.net/colipedia/index.php/Escherichia_coli BL21]-cells with 1µl of the PLasmid-DNA which has just been isolated.
Friday, 15.06.
- Transformation of [http://ecoliwiki.net/colipedia/index.php/Escherichia_coli BL21] worked well
- Tributyrinagar plates with 1mM IPTG were generated, to induce the Lac-Promotor of pET16b, and to demonstrate the esterase-activity by the lysis of Tributyrin.
- afterwards Tributyrinagar plates are inoculated with cells which were transformed by Miniprep No.3,4,15,16 on Wednesday, 13.06.
- incubation: 37°C, 2 days
Week9/CW25
Monday, 18.06.
- Tributyrinagar plates are covered with cells, but no lysis is visible.
- plates are incubated for another day at 25°C.
Tuesday, 19.06.
- no lysis, plates stay incubated at 25°C.
Wednesday, 20.06.
- no lysis
Friday, 22.06.
- The plasmids No.3,4 (EstA x FSC x phoA x pET16b) and No.15,16 (EstA x Est13 x phoA x pET16b) are prepared for sequencing.
- Primer:
- (EstA x FSC x phoA x pET16b):
- T7 up
- T7 lo
- SOE EstA mut up lo
- [[SOE EstA mut up lo
- (EstA x Est13 x phoA x pET16b):
- T7 up
- T7 lo
- SOE Est13 mut up lo
- SOE Est13 mut up lo
Sequencing was not successfull.
It was discovered later, that there was a contamination of the DH5alpha cells with other plasmid-DNA, probably caused by Electroporation.
SOE-PCR
Due to this fact, the isolated plasmids were always mixed up with other plasmids, and sequencing had to fail.
The contamination of the cells may have also caused false-positive results of the colony-PCRs.
We suggest that the cutinase FSC has an additional lipase-activity, which causes a lysis of the cell-membrane and may have an selective effect on FSC-negative mutants while cultivating the cells.
BioBricks
BioBricks
Week 1 / CW 21
- PCR of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808028 PhoA], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808025 FsC] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808026 Est13]: 3 assays à 50 µl TAnneal = 57°C @ 20s
- Agarose gel electrohporesis of the PCR products yields no result due to too short annealing time
- PCR of PhoA, FsC and Est13: 3 assays à 50 µl TAnneal = 57/62/67°C @ 120s
- 1% Agarose gel electrohporesis and PCR clean-up using the Wizard SV Gel and PCR Clean-Up System
- M Est13 PhoA+RBSx2 PhoAx3 PhoA+RBS Est13x2 FsCx3
- Note: Accidental exchange of PhoA+RBS (57°C) and Est13 (57°C)
Week 2 / CW 22
- PCR of FsC: 3 assays à 50 µl TAnneal = 57/58,6/60°C @ 120s
- PCR of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808027 EstA] and Est13: 2 assays à 50 µl TAnneal = 57/62°C @ 120s
- Electroporation of electrocompetent DH5alpha @ 2,5kV/2mm cuvette using 1µl of [http://partsregistry.org/Part:pSB1C3 pSB1C3] insert cultivation on chlor LB plates overnight at 37°C -> no colonies -> growth over weekend at ~25°C
Week 3 / CW 23
- Purity Plate of [http://partsregistry.org/Part:pSB1C3 pSB1C3] cultures 1d at 37°C
- DYT culture of pSB1C3 DH5alpha at 500 Hz / 37°C overnight
- Miniprep of the pSB1C3 cutures and determination of the concentration via NanoDrop yields of ~120ng/µl at 100µl
- Restriction digest of PhoA, EstA, Est13 and FsC (200 ng each) with PstI & EcoRI (à 20 µl)
- Clean-up of the restriction digest via ammonium sulfate precipitation utterly fails
Week 4 / CW 24
- Restriction digest of PhoA, EstA, Est13 and FsC (200 ng each) with PstI & EcoRI (à 20 µl)
- Clean-up of the restriction digest via Agarose gel electrohporesis and PCR clean-up using the Wizard SV Gel and PCR Clean-Up System] -> only FsC and Vector
- Pouring plates with Cmp 50x
- 1:3 and 1:5 Ligation of [http://partsregistry.org/Part:pSB1C3 pSB1C3] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808025 FsC] at 25°C overnight
- Electroporation of electrocompetent DH5alpha @ 2,5kV/2mm cuvette using 5µl of pSB1C3-FsC insert; cultivation on Cmp LB plates overnight at 37°C
Week 5 / CW 25
- Colony PCR and PCR of EstA, Est13 and PhoA
- Agarose gel electrohporesis of the PCR products
- PCR of EstA, Est13 and PhoA (Templates: PhoA = pEst100; EstA,Est13 = komp. Est13 mutated): 3 assays à 50 µl TAnneal = 57/62°C @ 120s
- 1% Agarose gel electrohporesis and PCR clean-up using the Wizard SV Gel and PCR Clean-Up System
Week 6 / CW 26
- Cultivation of [http://partsregistry.org/Part:pSB1C3 pSB1C3] in DYT (3ml) @ 37°C
- Miniprep of the pSB1C3 cutures and determination of the concentration via NanoDrop yields of ~47,02/52,11 ng/ml @ 30 µl each
- Restriction digest of EstA, Est13, PhoA and pSB1C3 (200 ng each) with PstI & EcoRI (à 20 µl) @ 37°C for 1:55h
- Heat inactivation of the restriction digest @ 55°C for 25min
Week 7 / CW 27
- Gelbild
Week 8 / CW 28
- Gelbild II
Week 9 / CW 29
Tuesday, 17.07.12
- each PCR is performed in 5 batches à 50 µL.
- TA = 57°C, tA = 30 s, tE = 2 min
- PCR on pEST100
- gene of interest: [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808028 PhoA] for designing part [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808028 BBa_K808028]
- primers: BBa PhoA up & BBa PhoA down
- PCR on pET26b(+)
- gene of interest: pNB-Est13 part1 for designing part BBa_K808026 via SOE PCR
- primers: BBa Est13 up & SOE Est13 mut lo
- PCR on pET26b(+)
- gene of interest: pNB-Est13 part2 for designing part BBa_K808026 via SOE PCR
- primers: BBa Est13 down & SOE Est13 mut up
- Agarose gel electrophoresis for quality control
- preparative Agarose gel electrophoresis
- Gel extraction of PCR products
- 1. PCR (gene of interest: phoA): c(PhoA)=19 ng/µL
- 2. PCR (gene of interest: pNB-Est13 part1): c(pNB-Est13 part1)=39 ng/µL
- 3. PCR (gene of interest: pNB-Est13 part2): c(pNB-Est13 part2)=40 ng/µL
- SOE PCR
- gene of interest: pNB-Est13 for designing part BBa_K808028
- primers: BBa Est13 up & BBa Est13 down
- Annotation: SOE PCR is performed in 5 batches à 50 µL
- TA1 = 52°C, tA1 = 25 s, tE1 = 75 s
- TA2 = 57°C, tA2 = 30 s, tE2 = 2 min
- stored over night at 10°C
Wednesday, 18.07.12
- qualitative Agarose gel electrophoresis of SOE PCR (from Tuesday, 17.07.12)
- preperative Agarose gel electrophoresis
- Gel extraction of SOE PCR produkt
- PCR 1
- gene of interest: EstA from SKV Date for designing part BBa_K808027
- primers: BBa EstA up and BBa EstA down
- Annotation: PCR is performed in 5 batches à 50 µL
- TA = 57°C, tA = 25 s, tE = 75 s
- GC Buffer is used
- preparative Agarose gel electrophoresis
- Gel extraction of PCR product
- gene of interest: EstA: c(pNB-Est13 part2)=119 ng/µL
- stored in freezer
Thursday, 19.07.12
- inoculation of 5 mL DYT-media with 5 µL Cmp and DH5alpha carrying pSB1C3 with part BBa_K808025 (FsC)
- DNA Digestion of the following parts
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808028 PhoA] from Tuesday, 17.07.12
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808026 pNB-Est13] fromTuesday, 17.07.12
- [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808027 EstA] from Wednesday, 18.07.12
- [http://partsregistry.org/Part:pSB1C3 pSB1C3] 1
- pSB1C3 2
- enzymes used: EcorI-HF & PstI-HF / for pSB1C3 2 SpeI is used instead of PstI
- in NEBuffer 4
- Digestion time: 2 h
- adding of Dephosphatase Antarctica from NEB on digested pSB1C3 1 & 2
- Ligation into digested and 5' dephosphorylated pSB1C3 1
- pNB-Est13
- PhoA
- EstA
- bacterial transformation by Electroporation of DH5alpha
Friday, 20.07.12
- the following Electroporation from Thursday, 19.07.12 worked
- inoculation of 5 mL DYT-media with 5 µL Cmp and two positive colonies from each transformation (from pNB-Est13 3 colonies are taken) .
- in addition FsC is already on plate, but to controle it, 2 colony PCRs are performed
- Ligation of EstA into digested and 5' dephosphorylated [http://partsregistry.org/Part:pSB1C3 pSB1C3]
- Electroporation of DH5alpha
- Miniprep of inoculated FsC in pSB1C3 (BBa_K808025) from Thursday, 19.07.12 failed
- inoculation of 5 mL DYT-media with 5 µL Cmp and DH5alpha carrying pSB1C3 with part BBa_K808025 (FsC)
- incubation at 37°C over weekend of transformations and picked colonies
Week 10 / CW 30
Monday, 23.07.12
- Colony PCR of picked colonies from Friday, 20.07.12
- 2-3 PhoA colonies, 4-5 FsC colonies, 6-8 pNB-Est13 colonies
- Miniprep of one inoculated colony in DYT-media with CAM from Friday, 20.07.12
- c(PhoA in [http://partsregistry.org/Part:pSB1C3 pSB1C3])=220 ng/µL
- c(pNB-Est13 in pSB1C3)=50 ng/µL
- inoculation of 2 x 5 mL DYT-media-Cmp with pSB1C3 carrying FsC
- second transformation of EstA from Friday, 20.07.12 worked, but too many colonies!
- purity plate is done
Tuesday, 24.07.12
- Miniprep of the 2 FsC DYT-media-Cmp cultures with [http://partsregistry.org/Part:pSB1C3 pSB1C3] carrying FsC from yesterday
- c(FsC in pSB1C3)=168 ng/µL
- purity plate of EstA in pSB1c3 is positive
- colonies picked
- Colony PCR with positive control by amplfifying an EstA from SKV
- Annotation: TA = 57°C, tA = 25 s, tE = 2 min, done with NEB- Phusion so its similiar to PCR 1
- GC Buffer is used
Wednesday, 25.07.12
- Agarose gel electrophoresis of Colony PCR
- worked out well
120725 Colony BBaEstA positiv probe auf SKV EstA.jpg
Thursday, 26.07.12
- Sequencing is ordered. The following premixes are used :
- 10 µL of c(PhoA in pSB1C3)=220 ng/µL, 1µL VR, 9 µL ddH2O
- 10 µL of c(PhoA in pSB1C3)=220 ng/µL, 1µL VF2, 9 µL ddH2O
- 7 µL of c(FsC in pSB1C3)=168 ng/µL, 1µL VR, 7 µL ddH2O
- 7 µL of c(FsC in pSB1C3)=168 ng/µL, 1µL VF2, 7 µL ddH2O
- 5 µL of c(EstA in pSB1C3)=350 ng/µL, 1µL VR, 9 µL ddH2O
- 5 µL of c(EstA in pSB1C3)=350 ng/µL, 1µL VF2, 9 µL ddH2O
- 5 µL of c(pNB-Est13 in pSB1C3)=50 ng/µL, 1µL VR, 9 µL ddH2O
- 5 µL of c(pNB-Est13 in pSB1C3)=50 ng/µL, 1µL VF2, 9 µL ddH2O
- in order to prepare DMSO stocks and Minipreps 5 mL DYT-media-CAM are inoculated with following colonies containing
Friday, 27.07.12
- inoculated pNB-Est13 in pSB1C3 from Thursday, 26.07.12 has not grown over night
- new colony pcked from plate and incubated in 5 mL DYT-media-CAM at 37°C over night
- Miniprep of 3 mL from the following over night DYT-media cultures from Thursday, 26.07.12
Week 11 / CW 31
Monday, 30.07.12
- bacterial transformation by Electroporation of DH5alpha with c(pNB-Est13 in pSB1C3)=50 ng/µL from Monday, 23.07.12
Tuesday, 31.07.12
- no results in sequencing our BioBricks / parts from Thursday, 26.07.12 except for FsC in pSB1C3
- new Colony PCR(similar to PCR 2) is done on
- no signal on qualitative Agarose gel electrophoresis
- Trouble shooting (see discussion below)
Trouble shooting
- Symptoms
- very much transformants on crossed out plates after bacterial transformation by Electroporation
- at first Colony PCR is positive
- Minipreps result in very high yields (exceeding >200 ng/µL)
- second Colony PCR is negative
- sequencing failed
- BUT FsC in pSB1C3
- has had not very much transformants
- has had a seuqencing result
- its Minipreps resulted in medium concentrations, settling around 90 ng/µL
- Diagnosis:
- our bacterial transformation is done by Electroporation
- therefor we use electroporation cuvettes, stored in DNA precipitating liquids like isopropanol or ethanol after being washed out.
- long before we started these electroporation cuvettes were in use.
- if not washed out carefully, DNA debris (like vectors and Inserts) accumulates in these storage liquids, due to precipitation
- when an Electroporation is performed with one of these cuvettes, probably containing only little DNA debris, the cells get transformed with numbers of different precipitated plasmids
- these plasmids contain a variety of resistances, reaching from CAM over KAN to Amp, allowing even "wrong" transformants to grow on selective media
- after a short period of time, bacterial colonies start to seperate and repel their plasmids, selecting only the most "comfortables"
- probably our plasmids, containing big genes on ligated vectors, are pushed out during this phase of selection
- this slow decrease of plasmid could explain the missing second positive signal of Colony PCR
- Strategy
- switching our method from Electroporation to Heatshock transformation
Wednesday, 01.08.12
- due to change of strategy the following PCR 1s are performed
- PCR on pEST100
- gene of interest: PhoA BioBrick for assembly of part BBa_K808028
- primers: BBa PhoA up & BBa PhoA down
- PCR on pEST100
- gene of interest: EstA part1 for SOE PCR of EstA (BBa_K808027)
- primers: BBa Est A up & SOE EstA mut PstI out lo
- PCR on pEST100
- gene of interest: EstA part2 for SOE PCR of EstA (BBa_K808027)
- primers: BBa EstA mut up & BBa EstA down
- PCR on pET26b(+)
- gene of interest: pNB-Est13 part1 for SOE PCR of pNB-Est13 (BBa_K808026)
- primers: BBa Est13 up & BBa Est13 mut lo
- PCR on pET26b(+)
- gene of interest: pNB-Est13 part2 for SOE PCR of pNB-Est13 (BBa_K808026)
- primers: BBa Est13 lo & BBa Est13 mut up
- Annotation: every PCR is performed in 3 batches à 50 µL
- TA = 57°C, tA = 25 s, tE = 75 s
- due to time pressure there will be no qualitative but only preperative Agarose gel electrophoresis
- PhoA only on analytical scale but on same Agarose gel electrophoresis with EstA part1
- Gel extraction of EstA part1, EstA part2, pNB-Est13 part1 and pNB-Est13 part2
- c(EstA part1)=19 ng/µL
- c(EstA part2)=110 ng/µL
- c(pNB-Est13 part1)=45 ng/µL
- c(pNB-Est13 part2)=48 ng/µL
- SOE PCRs
- SOE PCR
- gene of interest: EstA
- primers: EstA part1 & EstA part2
- SOE PCR
- gene of interest: pNB-Est13
- primers: pNB-Est13 part1 & pNB-Est13 part2
- as a control a PCR is performed on EstA part2
- Annotation: SOE PCR is performed in 3 batches à 50 µL
- TA1 = 52°C, tA1 = 30 s, tE1 = 75 s
- TA2 = 57°C, tA2 = 30 s, tE2 = 75 s
- stored over night at 10°C
- qualitative Agarose gel electrophoresis
- lane 2 pNB-Est13, lane 3 ESTA, lane 4EstA part2
- PCR Clean up by Promega Kit of PhoA, pNB-Est13, EstA
- DNA Digestion of the following parts
Thursday, 02.08.12
- Ligation into digested and 5' dephosphorylated pSB1C3
- pNB-Est13
- PhoA
- EstA
- Annotation: Ligation is performed in a 20 µL batch
- digested pSB1C3 still availabe from Thursday, 19.07.12
- 1:5 & 1:10 ratio
- incubation at room temperature for 30 min
- Heatschock transformation of DH5alpha with our ligation batches
- incubation over night at 37°C
Friday, 03.08.12
- transformation succeeded
- colonies picked to inoculate 5 ml DYT-media-CM
Saturday, 04.08.12
- Miniprep of picked colonies
- test DNA Digestion with EcoRI-HF and PstI-HF
- PCR 2 on preped plasmids with primers VR & VF2, and the respective BBa primers
- TA = 60°C, ta = 35s, tE = 25 s
- qualitative Agarose gel electrophoresis 1% agarose
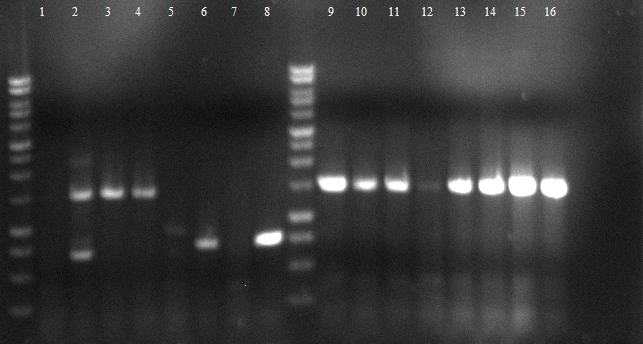
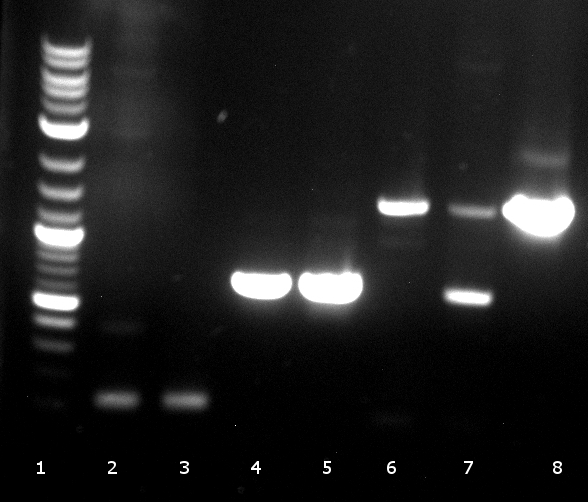
120804 Verdau PhoA, EstA, Est13, PCR auf EstA, Est 13, PhoA, Kontrolle auf Insert und Vektor.jpg
- digestion: 2 PhoA, 3 EstA, 4 pNB-Est13 PCR: 6 EstA with BBa primers, 7 EstA with VR & VF2, 8 pNB-Est13 with BBa primers, 10 pNB-Est13 with VR & VF2, 11 PhoA with BBa primers, 12 PhoA with VR & VF2
- tests succeeded
- Annotation: failure in mixing sequencing premixes
- retransformation of DH5alpha by Heatshock transformation with the following plasmids with 1 µL each
Week 12 / CW 32
Assembly of our composite part RBS-PhoA-His6tag-pNBEst13-Myctag-EstA (BBa_K808030) is reached by synthesis (from GENEART) of 2 different gene parts:
- bba prefix-PhoA-His6tag-pNBEst13-BsaI site
- BsaI-Myctag-EstA- bba suffix
- the RBS will be added by PCR with primers XbaI Rbs Phoa up (ignoring the bba prefix but adding an XbaI site upstream of the RBS) & Est13 Bsa1 lo, ignoring the bba prefix but adding an XbaI site upstream of the RBS and for the downstream part primers EstA Bsa1 up andBBa EstA down are used
- both products can be assembled by using BsaI. An restriction enzyme, with an restriction site in a defined distance to its recognition site
Monday, 06.08.12
- Heatshock transformation of DH5alpha with 1 µL synthesis products (c=100 ng/µL):
- DNA Digestion of 10 µL = 1 µg of both synthesis products
- bba prefix-PhoA-His6tag-pNBEst13-BsaI site digested with EcoRI-HF and BsaI-HF in a 50 µL batch
- BsaI-Myctag-EstA- bba suffix digested with BsaI-HF and PstI-HF in a 50 µL batch
- preparative Agarose gel electrophoresis
- Failure due to overlooked additional PstI site in GENEART Vectors
- inoculation of 5 ml DYT-media-CAM with retransformations from Saturday, 04.08.12
- incubation at 37°C over night
Tuesday, 07.08.12
- Colony PCR on 4 colonies of each transformation of synthesis product from Monday, 06.08.12
- Annotation: PCR is done with house-taq, similar to PCR 2
- TA = 57°C, tA = 30 s, tE = 90 s
- Colony PCR succeeded
- from bba prefix-PhoA-His6tag-pNBEst13-BsaI site colonies 5, 2, 11 are picked
- from BsaI-Myctag-EstA- bba suffix colonies 1, 10, 16 are picked
- inoculation of 5 ml DYT-media-KAN with colonies
- incubation over night at 37°C
- inoculated 5 mL cultures from Monday, 06.08.12 are stored at 4°C
Wednesday, 08.08.12
- Miniprep of following 5 mL cultures
- from Monday, 06.08.12
- from Tuesday, 07.08.12
- bba prefix-PhoA-His6tag-pNBEst13-BsaI site
- colony 2 c=438 ng/µL
- colony 5 c=509 ng/µL
- colony 11 c=291 ng/µL
- BsaI-Myctag-EstA- bba suffix
- colony 1 c=381 ng/µL
- colony 10 c=420 ng/µL
- colony 16 c=263 ng/µL
- bba prefix-PhoA-His6tag-pNBEst13-BsaI site
- DNA Digestion
- of bba prefix-PhoA-His6tag-pNBEst13-BsaI site colony 11
- performed in a 40 µL batch
- BsaI and EcoRI are used
- digestion for 3 h
- of BsaI-Myctag-EstA- bba suffix
- performed in a 40 µL batch
- BsaI and SpeI are used
- digestion for 3 h
- of bba prefix-PhoA-His6tag-pNBEst13-BsaI site colony 11
- preperative Agarose gel electrophoresis
- 3-4 cut out bba prefix-PhoA-His6tag-pNBEst13-BsaI site, 5-6 cut out BsaI-Myctag-EstA- bba suffix
- Gel extraction
- c(bba prefix-PhoA-His6tag-pNBEst13-BsaI site)= 8 ng/µL
- c(BsaI-Myctag-EstA- bba suffix)= 26 ng/µL
- ligation of both inserts in pSB1C3 2 from Thursday, 19.07.12
- 1:5:5 ratio is performed in two 20 µL batches (one incubated at 4°C, the other at room temperature over night)
- new sequencing is ordered of the following plasmids
Thursday, 09.08.12
- Heatshock transformation of DH5alpha with 10 µL of each ligation batch from yesterday
Friday, 10.08.12
- transformation from yesterday suceeded
- one colony per plate (1:5:5 & 1:10:10 at room temperature and 1:5:5 & 1:10:10 at room 4°C)is picked for inoculation of 5 mL DYT-media-CAM
Saturday, 11.08.12
- Miniprep of picked colonies from yesterday
- testing ligation by DNA Digestion of plasmids with EcorI-HF & PstI-HF
- Colony PCR on picked colonies
- Ligation and transformation succeeded
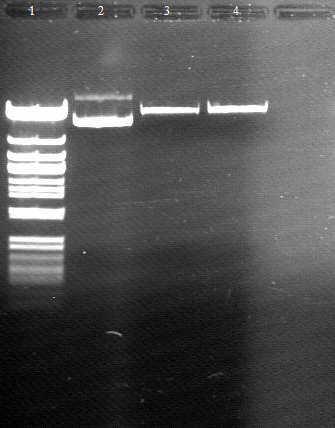
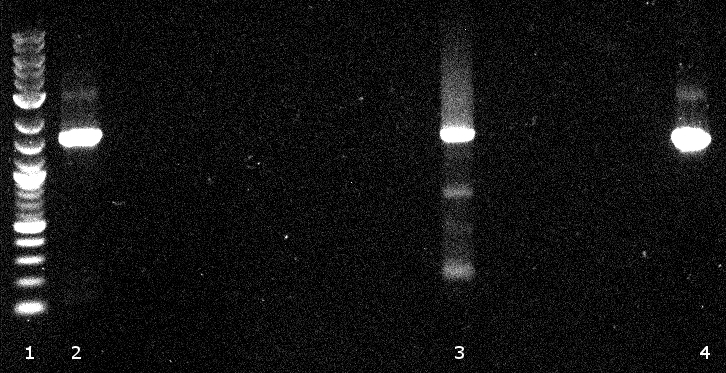
120811 PCR (VR,VF2) und Verdau (EcoRI,PstI) auf BBa Gesamt von Geneart.jpg
- 2-5 bba prefix-PhoA-His6tag-pNBEst13-Myctag-EstA- bba suffix running at round about 3 kb
Week 13 / CW 33
Monday, 13.08.12
- Preparation for designing part BBa_K808032 which is an arabinose regulated RBS-PhoA-His6tag-pNBEst13-Myctag-EstA
- in order to add RBS upstream of PhoA of bba prefix-PhoA-His6tag-pNBEst13-BsaI site the following PCRs are performed
- PCR 1 on bba prefix-PhoA-His6tag-pNBEst13-BsaI site, as a template the pure synthesis product is used
- TA = 57°C, tA = 25 s, tE = 2 min
- primers: XbaI Rbs Phoa up & Est13 Bsa1 lo
- PCR 1 on BsaI-Myctag-EstA- bba suffix, as a template the pure synthesis product is used
- TA = 57°C, tA = 25 s, tE = 2 min
- primers: EstA Bsa1 lo & BBa EstA down
- PCR 3 on preped colony (1:5:5 room temperature from Friday, 10.08.12)
- TA = 57°C, tA = 25 s, tE = 2 min
- primers: XbaI Rbs Phoa up & BBa EstA down
- Annotation: each PCR is performed in 4 batches à 50 µL
- 2 qualitative Agarose gel electrophoresis
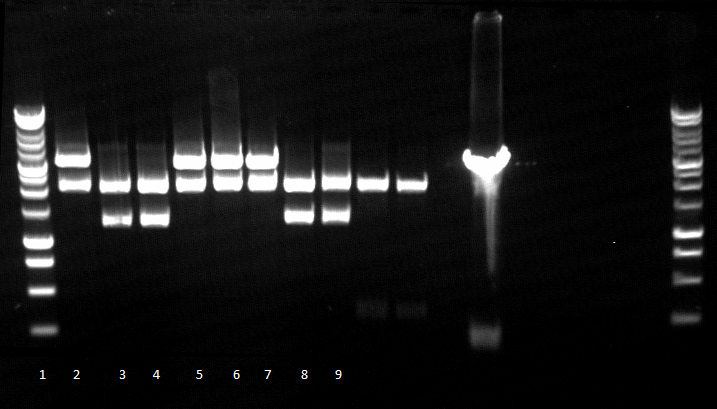
120813 PCR 1-4 RBSPE 5-8 EXN-Myc-Est 9-12.jpg
120813 PCR Q5 auf bba gesamt konstrukt.jpg
- 2-4 bba prefix-RBS-PhoA-His6tag-pNBEst13-Myctag-EstA-bba suffix
- PCRs succeeded
- PCR clean up of 1.-3. PCR
- DNA Digestion of pSB1C3 carrying RFP part ????
- DNA Digestion of part BBa_K808000 (pSB1C3 carrying Arabinose inducible promotor)
- preparative Agarose gel electrophoresis
- c(1.PCR)= 147 ng/µL
- c(2.PCR)= 133 ng/µL
- c(3.PCR)= 96 ng/µL
- c(cut S/P BBa_K808000: Arabinose inducible promotor) possibility 1= 14 ng/µL
- c(cut S/P BBa_K808000: Arabinose inducible promotor) possibility 2= 14 ng/µL
- c(cut X/P pSB1C3 carrying RFP)=20 ng/µL
- Ligation at 4°C over night of the following combinations:
- 1.PCR & 2.PCR in pSB1C3 carrying RFP
- 3.PCR in pSB1C3 carrying RFP
- 1.PCR & 2. PCR in BBa_K808000 possibility 1
- 3.PCR in BBa_K808000 possibility 1
- 1.PCR & 2. PCR in BBa_K808000 possibility 2
- 3.PCR in BBa_K808000 possibility 2
Tuesday, 14.08.12
- Heatshock transformation of ligation batches 1-6 from yesterday
Wednesday, 15.08.12
- transformation succeeded
- inoculation of 5 mL DYT-media-CAM of the following colonies
- colony 1.1
- colony 2.1
- colony 2.2
- colony 3.1
- colony 4.1
- colony 4.2
- colony 5.1
- colony 5.2
- colony 6.1
- colony 6.2
- incubation over night at 37°C
- Colony PCR on colonies 1.1-6.2 with house-taq
Thursday 16.08.12
- Miniprep of colonies from yesterday
- DNA Digestion of preped colonies with EcorI-HF & PstI-HF
- incubation for 1.5 h at 37°C
- qualitative Agarose gel electrophoresis
- FAILURE, because bands on gel are inconsistent
- 2 colony 1.1, 3 colony 2.1, 3 colony 2.2, 4 colony 3.1, 5 colony 4.1, 6 colony 4.2, 7 colony 5.1, 8 colony 5.2
- for further information see discussion below
- new inoculation of 5 mL DYT-media-CAM of Colonies 1.1 - 5.2
Trouble shooting
What should have been transformed:
- colony 1.1: 1.PCR & 2.PCR in pSB1C3
- should be RBS-PhoA-His6tag-pNBEst13-Myctag-EstA (BBa_K808030) in pSB1C3 (length: ~ 3.1 kb)
- colony 2.1: 3.PCR in pSB1C3 upstream of part BBa_K808000 (Arabinose inducible promotor) possibility 1
- should be Arabinose regulated RBS-PhoA-His6tag-pNBEst13-Myctag-EstA (BBa_K808032) in pSB1C3 (length: ~ 4.3kb)
- colony 2.2: similar to colony 2.1
- colony 3.1: 3.PCR in pSB1C3 upstream of part BBa_K808000 (Arabinose inducible promotor) possibility 2
- should be Arabinose regulated RBS-PhoA-His6tag-pNBEst13-Myctag-EstA (BBa_K808032) in pSB1C3 (length: ~ 4.3kb) like colony 2.1
- colony 4.1: 3.PCR in pSB1C3
- should be RBS-PhoA-His6tag-pNBEst13-Myctag-EstA (BBa_K808030) in pSB1C3 (length: ~ 3.1 kb)
- colony 4.2: similiar to colony 4.1
- colony 5.1: 1.PCR & 2.PCR in pSB1C3 upstream of part BBa_K808000 (Arabinose inducible promotor) possibility 1
- should be Arabinose regulated RBS-PhoA-His6tag-pNBEst13-Myctag-EstA (BBa_K808032) in pSB1C3 (length: ~ 4.3kb)
- colony 5.2: similar to colony 5.1
Conclusion:
- estimated gene length was aorund 4,3 kb (colony 5.1, 5.2, 2.1, 2.2 = part BBa_K808032) or 3,1 kb (colony 1.1, 4.1, 4.2 part BBa_K808030)
- digestion with EcoRI-HF & PstI-HF of all ligations from Thursday 16.08.12 in part BBa_K808000 (Arabinose inducible promotor) possibility 1 shows bands at round about 3 - 3.5 kb
- when digested with same enzymes of ligations into part BBa_K808000 (Arabinose inducible promotor) possibility 2, bands are shown at round about 1 - 1.5 kb
- this leads to our concluison that BBa_K808000 (Arabinose inducible promotor) possibility 1 was the not digested DNA band, because resulting gene length of cut insert correlates with length of BBa_K808000 (Arabinose inducible promotor)
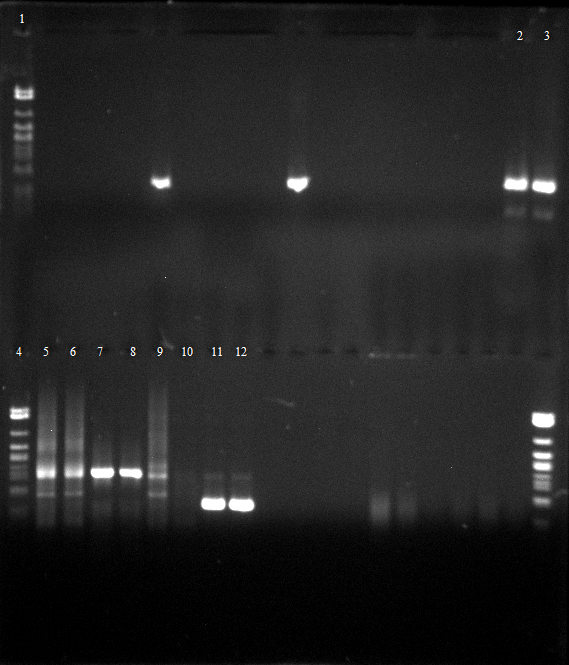
- for a sharper solution an Agarose gel electrophoresis with 0.5 % agarose is performed for digestions of colonies 1.1, 3.1, 4.1, 4.2 plus batches from Colony PCR from Wednesday, 15.08.12
- 2-5 colonies 1.1, 3.1, 4.1, 4.2, 6 - 13 colony 1.1, 2.1, 2.2, 3.1, 4.1, 4.2, 5.1, 5.2
Friday, 17.08.12
- the image above means that:
- colony 5.1, 5.2, 2.1, 2.2 are only carrying part [[BBa_K808000 which encodes for the arabinosis inducable promotor
- but colony 1.1, 2.1, 2.2 are carrying 1.PCR & 2.PCR or 3.PCR in pSB1C3
- Miniprep of named colonies in liquid DYT-culture from Thursday 16.08.12
- half culture is preped, half culture serves for 10% DMSO stocks
- c(1.1: BBa_K808030)= 81 ng/µL
- c(4.1: BBa_K808030)= 128 ng/µL
- c(4.2: BBa_K808030)= 79 ng/µL
- c(2.1: BBa_K808000)= 90 ng/µL
- c(2.2: BBa_K808000)= 78 ng/µL
- c(5.1: BBa_K808000)= 64 ng/µL
- c(5.2: BBa_K808000)= 62 ng/µL
- for BioBrick assembly we perform a DNA Digestion of
- BBa_K808000 (colony 1.1, 4.1, 4.2), with EcoRI / SpeI, serves as upstream part
- BBa_K808030 (colony 5.1, 5.2, 2.1, 2.2), with EcoRI / XbaI, serves as downstream part
- preperative Agarose gel electrophoresis
- Gel extraction
- downstream part
- c(1.1: BBa_K808030)= 21.5 ng/µL
- c(4.1: BBa_K808030)= 54 ng/µL
- c(4.2: BBa_K808030)= 58 ng/µL
- upstream part
- c(2.1: BBa_K808000)= 12 ng/µL
- c(2.2: BBa_K808000)= 11 ng/µL
- c(5.1: BBa_K808000)= 10 ng/µL
- c(5.2: BBa_K808000)= 5 ng/µL
- ligation of upstream infron of downstream part
- colony 5.1 (BBa_K808000) in colony 1.1 (BBa_K808030)
- colony 2.2 (BBa_K808000) in colony 4.2 (BBa_K808030)
- colony 2.1 (BBa_K808000) in colony 4.1 (BBa_K808030)
- inoculation of 5 mL DYT-media-CAM of the following colonies
- colony 1.1
- colony 2.1
- colony 2.2
- colony 3.1
- colony 4.1
- colony 4.2
- colony 5.1
Week 14 / CW 34
Monday 20.08.12
- Miniprep of incoucaltes colonies from Friday, 17.08.12
- c(1.1: BBa_K808030)= 54 ng/µL
- c(4.1: BBa_K808030)= 74 ng/µL
- c(4.2: BBa_K808030)= 60 ng/µL
- c(2.1: BBa_K808000)= 47 ng/µL
- c(2.2: BBa_K808000)= 42 ng/µL
- c(5.1: BBa_K808000)= 56 ng/µL
- sequencing of
- colony 1.1 using VR & VF2, 1.5 µL each
- colony 5.1 using VR, SOE EstA mut lo, SOE EstA mut up & VF2, 1.5 µL each
Tuesday 21.08.12
- Ligation of BBa_K808000 upstream of BBa_K808032 in pSB1C3 ( to design BBa_K808032) from Friday, 17.08.12 worked well
- 6 colonies are picked in order to perform a Colony PCR and for inoculation of 5 mL DYT-media-CAM
- colony A 4.1/2.1 ligated at 4°C
- colony B 4.1/2.1 ligated at 37°C
- colony C 1.1/5.1 ligated at 4°C
- colony D 1.1/5.1 ligated at 37°c
- colony E 4.2/2.2 ligated at 4°C
- colony F 4.2/2.2 ligated at 37°C
- Colony PCR
- Agarose gel electrophoresis
- did not work due to immense gene legth ( around 4 kb)
- Restriction digest for testing is planned
- inoculation of 5 mL DYT-media-CAM with colony E,F,C,D
Wednesday, 22.08.12
- Miniprep of inoculated 5 mL DYT-media-CAM with colony E,F,C,D
- c(colony C 1.1/5.1 ligated at 4°C)=46 ng/µL
- c(colony D 1.1/5.1 ligated at 37°c)=46 ng/µL
- c(colony E 4.2/2.2 ligated at 4°C)=60 ng/µL
- c(colony F 4.2/2.2 ligated at 37°C)=58 ng/µL
- DNA Digestion of 15 µL of preped plasmids with EcoRI-HF & PstI-HF
- expected insert length: 4.4 kb (BBa_K808000+BBa_K808030)
- Agarose gel electrophoresis with 0.8% agarose
- ligation worked well, bands are in estimated length
- lane 2-5: colony 4.2/2.2, lane 6-9: colony 5.1/1.1
- due to rather bad results of Miniprep colonies are used for inoculation of 5 mL DYT-media-CAM again
- incubation over night at 37°C
- evaluation of sequencing from Wednesday, 08.08.12
- PhoA did not work
- EstA is sequenced inconsistent
- pNB-Est13 still got its illegal PstI-site
- but at leat FsC is sequenced completely
- new sequencing of
- PhoA in pSB1C3, primers VF2 & VR, 1.5 µL each
- EstA in pSB1C3, primers SOE EstA mut up & BBa EstA down, 1.5 µL each
- PCR 1 performed on BBa_K808032(preped colony C 1.1/5.1 ligated at 4°C)
- TA = 57°C, tA = 25 s, tE = 2 min
- primers BBa Est13 up & BBa Est13 down
- performed in 3 batches à 50 µL
- analytical Agarose gel electrophoresis 2% agarose
- [PCR Clean up]]
- DNA Digestion of PCR product with EcoRI-HF & PstI-HF over night at 37°C
Thursday, 23.09.12
- Miniprep of colonies from
- c(colony C 1.1/5.1 ligated at 4°C)=147 ng/µL
- c(colony D 1.1/5.1 ligated at 37°c)=131ng/µL
- c(colony E 4.2/2.2 ligated at 4°C)=85 ng/µL
- c(colony F 4.2/2.2 ligated at 37°C)=128 ng/µL
- preparative Agarose gel electrophoresis of digested pNB-Est13 from yesterday
- Gel extraction leads to c(pNB-Est13- cut E/P)=25 ng/µL
- Ligation of digested pNB-Est13 in digested pSB1C3
- 1:5 ratio is used
- Heatshock transformation of DH5alpha with 10 µL of ligation batch
- incubation of transformed cells at 37°C over night
Friday, 24.08.12
- transformation of ligation from yesterday succeeded, colony picked and inoculation of 5 mL DYT-media-CAM
- sequencing of
- BBa_K808032 (colony C 1.1/5.1 ligated at 4°C), primers VF2, XbaI Rbs Phoa up, BBa Ara Promo lo, SE Est13 mut up, SOE Est13 mut lo, BBa EstA down, VR, 1.5 µL each,
Week 15 / CW 35
Monday, 27.08.12
- Miniprep of picked colony from [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808026 BBa_K808026] ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K808026 pNB-Est13] in [http://partsregistry.org/Part:pSB1C3 pSB1C3])
- c([http://partsregistry.org/wiki/index.php?title=Part:BBa_K808026 pNB-Est13] in [http://partsregistry.org/Part:pSB1C3 pSB1C3])=116 ng/µL
- sequencing of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808026 pNB-Est13] in [http://partsregistry.org/Part:pSB1C3 pSB1C3]
- using primers VF2, VR, SOE Est13 mut up, SOE Est13 mut lo
- test expression of BBa_K808032 (colony C 1.1/5.1 ligated at 4°C) in DH5alpha
- BBa_K808032 is an arabinose regulated RBS-PhoA-His6tag-pNBEst13-Myctag-EstA
- inoculation of 50 mL DYT-media-CAM with DH5alpha carrying [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808032 BBa_K808032]
- stop incubation at at OD600=0.7
- storaging cultures on ice for 15mins
- inducing with ~ 1.5% L-arabinose (using 3.75 mL of 20% mw L-arobinose solution)
- incubation at 20°C, 25°C and 30°C over night
Tuesday, 28.08.12
- antibody staining
- detection of surface expression of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808032 BBa_K808032] by using FACS
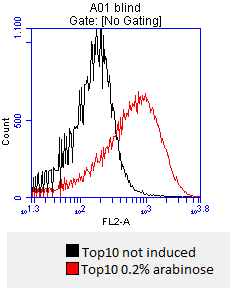
- positive signal increases with higher temperature, staining succeeded
- protein purification of test expression without running IMAC afterwards, just using supernatant and cell debris pellet
- 32 µL of supernatant is used
- 3 mL pellet of each testexpression is treated with more SDS as a detergent, in order to solve membrane proteins
- running two SDS-tris gels with an myc positive probe
- one of these gels is used for performing a Westernblot
- using mouse-antimyc antibody as first antibody and goat-antimouse-AP as second antibody
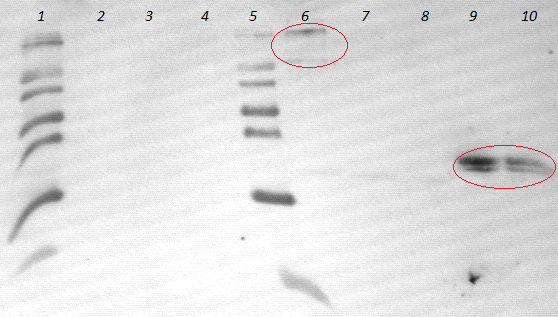
- lane 2-4: supernatant of induced and disrupted cells. Level of induction decreases from lane 2 (1.5% arabinose) to lane 4 (0.5% arabinose)
- lane 6-8: Pellet of induced and disrupted cells, treated with high SDS concentrations. Level of induction decreases from lane 6 (1.5% arabinose) to lane 8 (0.5% arabinose).
- Lane 9-10 show myc positive probes.
- The blot shows that our expression worked of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808032 BBa_K808032] and proofs the membrane intercalation of our chimeric protein [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808032 BBa_K808030] because the blot only gives positive signals when high concentrations of detergent where used to treat the cell debris pellet.
- both are not very good in their solution, we do it again with Laemmli gels
Wednesday, 29.08.12
- evalutation of sequencing from last week:
- PhoA worked
- EstA worked
- pNB-Est13 worked
- RBS-PhoA-His6tag-pNBEst13-Myctag-EstA worked
- arabinose regulated RBS-PhoA-His6tag-pNBEst13-Myctag-EstA
Thursday, 30.08.12
- running 2 SDS-Laemmli gels
- pellet is treated with n-Dodecyl-ß-maltoside (DDM) in order to solve membrane proteins
- one gel is used to perform a second Western blot with a myc positive probe
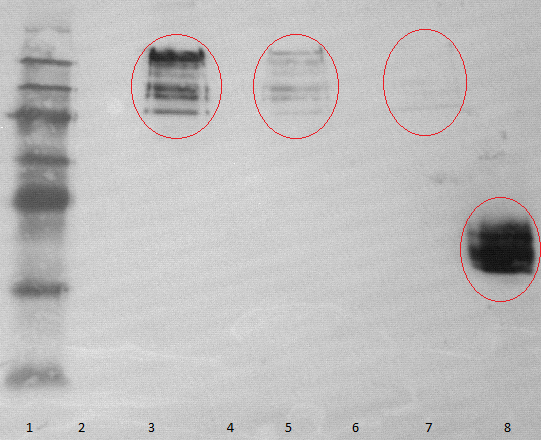
- 2 supernatant 30°C, 3 pellet (treated with detergent) 30°C, 4 supernatant 25°C, 5 pellet (treated with detergent) 25°C, 6 supernatant 20°C, 7 pellet (treated with detergent) 20°C, 8 myc positive probe
- as we can see, only the treated 120831 Westermblot 2 auf Gesamt konstrukt aus DH5 alpha 5111pellets show positive signals when treated with antibodies, so we have expressed our membrane protein [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808032 BBa_K808032]
- now our lab can start with quantification of enzymatic activites of BBa_K808032 on PET or model substrates
Activity tests of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808032 BBa_K808032]
We were able to generate our BioBricks [http://partsregistry.org/Part:BBa_K808030 BBa_K808030] (chimeric, membrane bound, hydrolytic protein) and [http://partsregistry.org/Part:BBa_K808032 BBa_K808032] ( arabinose inducible operon, regulating the expression of BBa_K808030). Funcionality of both parts will be described by a bunch of tests:
- SDS PAGE
- Western blot
- [http://en.wikipedia.org/wiki/Flow_cytometry#Labels flow cytometry] (after antibody staining)
- screening for hydrolysis by bacterial colonies using Tributyrin agar plates
- pNP-Assay with living cells using para-Nitrophenylbutyrate
- As hosts we will use E.coli strains [http://ecoliwiki.net/colipedia/index.php/TOP10 Top10], [http://ecoliwiki.net/colipedia/index.php/DH5_alpha DH5alpha] or [http://ecoliwiki.net/colipedia/index.php/MG1655 Mg1655]
Week 1 / CW 35
Friday, 31.08.12
- Activity assay in DYT with DH5α containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808032 BBa_K808032] (arabinose inducable RBS-PhoA-His6tag-pNBEst13-Myctag-EstA)
| Component | test tube 1 | test tube 2 | test tube 3 | test tube 4 |
|---|---|---|---|---|
| DYT-medium | 8 mL | 8 mL | 8 mL | 8 mL |
| PET particle | yes | yes | yes | yes |
| bacteria | yes | yes | yes | no |
| induced | 1.5% L-arabinose | 1.5% L-arabinose | no | no |
- test seemed to have worked: but an induced test tube without PET-granula was missing
Week 2 / CW 36
Tuesday, 04.09.12
- Activity assay in DYT with DH5α containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808032 BBa_K808032]
| Component | test tube 1 | test tube 2 | test tube 3 | test tube 4 | test tube 5 |
|---|---|---|---|---|---|
| DYT-medium | 8 mL | 8 mL | 8 mL | 8 mL | 8 mL |
| PET particle | yes | yes | no | yes | yes |
| bacteria | yes | yes | yes | no | yes |
| induced | 1.5% L-arabinose | 1.5% L-arabinose | 1.5% L-arabinose | no | no |
- test tube 3: DYT without bacteria contains CAM, Kan, AMP
- looks good but test tube 2 shows no significant change of colour
Wednesday, 05.09.12
- Activity assay in DYT with DH5α containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808032 BBa_K808032]
| Component | tube 1 | tube 2 | tube 3 | tube 4 | tube 5 | tube 6 | tube 7 | tube 8 | tube 9 |
|---|---|---|---|---|---|---|---|---|---|
| DYT-medium | 8 mL | 8 mL | 8 mL | 8 mL | 8 mL | 8 mL | 8 mL | 8 mL | 8 mL |
| PET particle | yes | yes | yes | yes | no | no | yes | yes | yes |
| bacteria | yes | yes | yes | yes | yes | yes | yes | yes | no |
| induced | 1.5% L-arabinose | 1.5% L-arabinose | 1.5% L-arabinose | 1.5% L-arabinose | 1.5% L-arabinose | 1.5% L-arabinose | no | no | no |
- test tube 9: DYT without bacteria contains CAM, Kan, AMP
- all induced tubes turned yellow, even without PET-granula as a substrate
- no more avtivity tests, we are awaiting the evaluation of test expression series of the [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 arabinose inducible promoter] conjugated to GFP
Trouble shooting
- evaluation shows a very high sensisty of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808000 arabinose inducible promoter [BBa_K808000]] even to low concentrations of L-arabinose (expression starts at around 0.01%)
- induction of the DH5α with 1.5% arabinose ended fatal due to extrem expression of the transmembrane construct [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808030 RBS-PhoA-His6tag-pNBEst13-Myctag-EstA]
- starting test expressions with lower L-arabinose concentrations ranging from 0.05% - 1%
Thursday, 06.09.12
- Activity assay in DYT with DH5α containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808032 [BBa_K808032]] (arabinose inducable RBS-PhoA-His6tag-pNBEst13-Myctag-EstA)
| Component | tube 1 | tube 2 | tube 3 | tube 4 | tube 5 | tube 6 | tube 7 | tube 8 | tube 9 | tube 10 | tube 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| DYT-medium | 5 mL | 5 mL | 5 mL | 5 mL | 5 mL | 5 mL | 5 mL | 5 mL | 5 mL | 5 mL | 5 mL |
| PET particle | yes | yes | yes | yes | yes | yes | no | no | no | no | no |
| bacteria | yes | yes | yes | yes | yes | yes | yes | yes | yes | yes | no |
| induced | 1% L-arabinose | 1% L-arabinose | 0.75% L-arabinose | 0.75% L-arabinose | no | no | 1% L-arabinose | 1% L-arabinose | 0.75% L-arabinose | 0.75% L-arabinose | no |
- test tube 11: DYT without bacteria contains CAM, Kan, AMP
- all induced tubes turned yellow, even without PET-granula as a substrate
Week 3 / CW 37
we stopped the trial of quantification via PET-substrates because in terms of color the change of pH value (due to acid groups that are released by degradtion) is not very exactly. In order to get reproducible data, we are using other activity assays:
- pNP-Assay
- activity assay on LB-Tributyrin-CAM-plates used for screening of hydrolytic bacterial colonies.
Monday, 10.09.12
- Heatshock transformation of [http://ecoliwiki.net/colipedia/index.php/MG1655 Mg1655] with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808032 BBa_K808032] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808030 BBa_K808030]
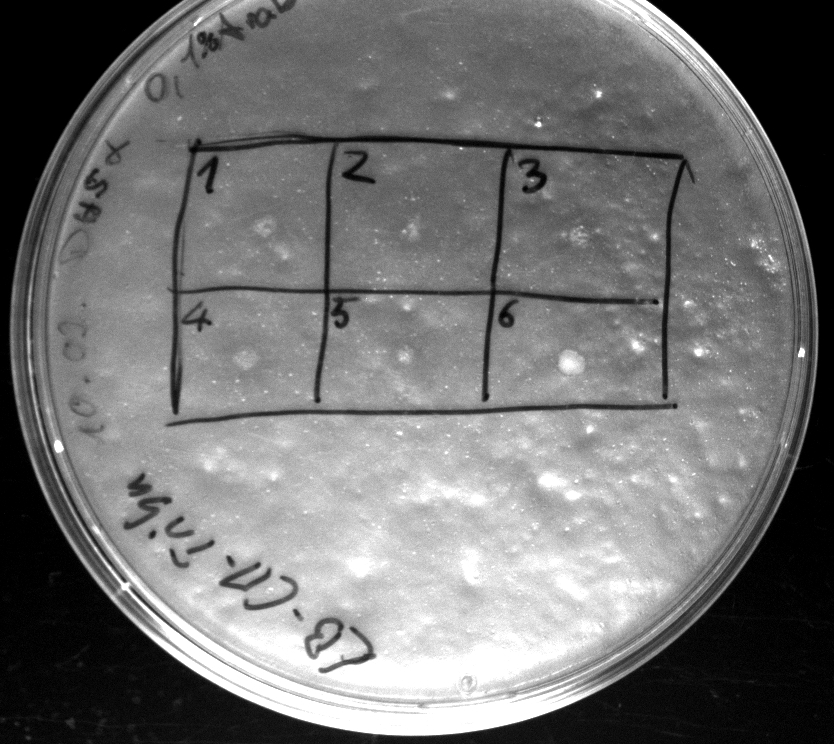
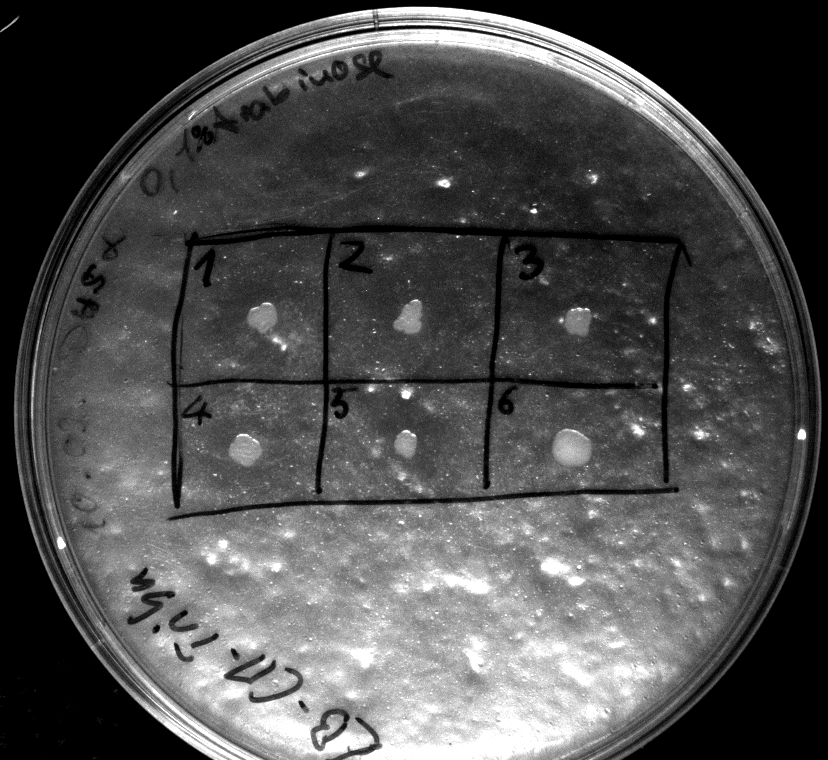
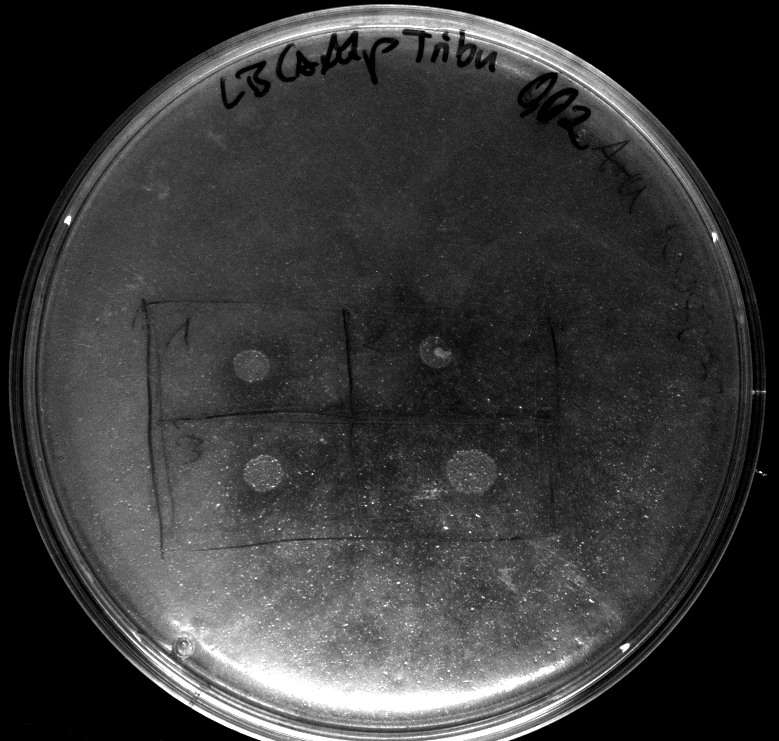
- activity assay on LB-Tributyrin-CAM-plates with L-arabinose concentrations: 0.1%, 0.2%, 0.4%
- colony 1-5 are [http://ecoliwiki.net/colipedia/index.php/DH5_alpha DH5 alpha] containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808032 BBa_K808032] but colony 6 is just DH5 alpha carrying [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808030 BBa_K808030]
- incubated over night at 37°C
Tuesday, 11.09.12
- inoculation of 5 mL DYT-media-CAM with K1 and K2 of transformed [http://ecoliwiki.net/colipedia/index.php/MG1655 Mg1655] from yesterday
- acticity tests on LB-Tributyrin-CAM-plates shows good results
- from now on incubation at room temperature
Wednesday, 12.09.12
- Plasmid preparation of inoculated [http://ecoliwiki.net/colipedia/index.php/MG1655 Mg1655] colonies K1 and K2
- DNA digestion with EcoRI-HF & PstI-HF
- qualitative AGE
Thursday, 13.09.12
- the activity assay on LB-Tributyrin-CAM-plates from tuesday gives good results
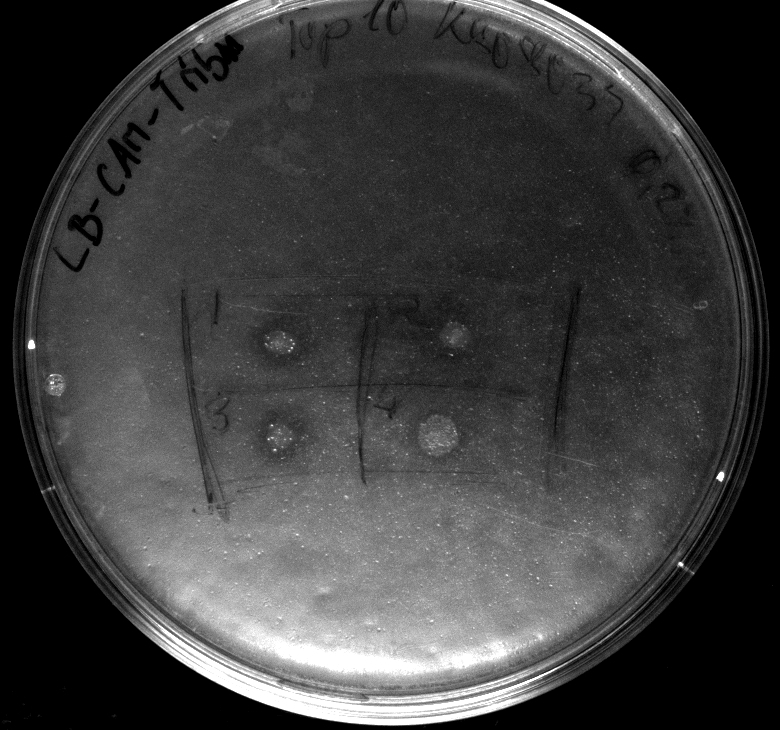
- to perform a better expression of [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808030 BBa_K808030] we will use [http://ecoliwiki.net/colipedia/index.php/TOP10 Top10] as a host for [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808032 BBa_K808032]. Its unability to metabolize L-arabinose is crucial for our decision. We hope to generate good expression levels.
- Electroporation (after washing ecletroporation cuvettes very carefully) of [http://ecoliwiki.net/colipedia/index.php/TOP10 Top10] with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808032 BBa_K808032]
Friday, 14.09.12
- yesterdays electroporation worked well
- inoculation of 50 mL DYT-media-CAM with colony from plate of [http://ecoliwiki.net/colipedia/index.php/TOP10 Top10] carrying [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808032 BBa_K808032]
Saturday, 15.09.12
- making 10% DMSO stocks from 50 mL [http://ecoliwiki.net/colipedia/index.php/TOP10 Top10] culture
Week 4/ KW 38
Monday, 17.09.12
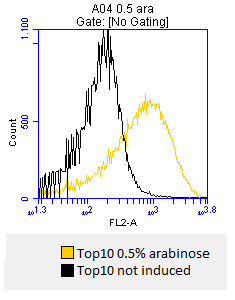
- inoculation of 4 x 50 mL DYT-media-CAM with [http://ecoliwiki.net/colipedia/index.php/TOP10 Top10] stocks
- at OD600=0,5 the cultures are incduced with:
- 0.02% mw L-arabinose
- 0.2% mw L-arabinose
- 0.5% mw L-arabinose
- one culture is not induced and will serve as a negative control
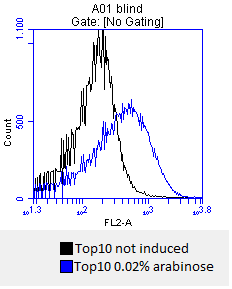
- after 3 hours an antibody staining is performed on our expressed [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808032 BBa_K808032] containing a myc-epitope
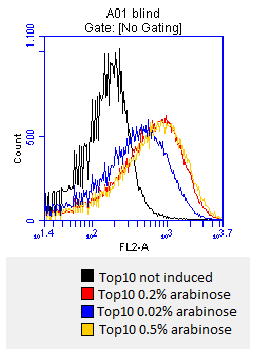
- checking surface expression levels via [http://en.wikipedia.org/wiki/Flow_cytometry#Labels flow cytometry]
- inducing worked well, we can go on with screening the enzymatic activity
Tuesday, 18.09.12
- Electroporation of [http://ecoliwiki.net/colipedia/index.php/TOP10 Top10] with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808030 BBa_K808030], serving as a negative (directly used after 1h of incubation at 37°C in pure DYT-medium)
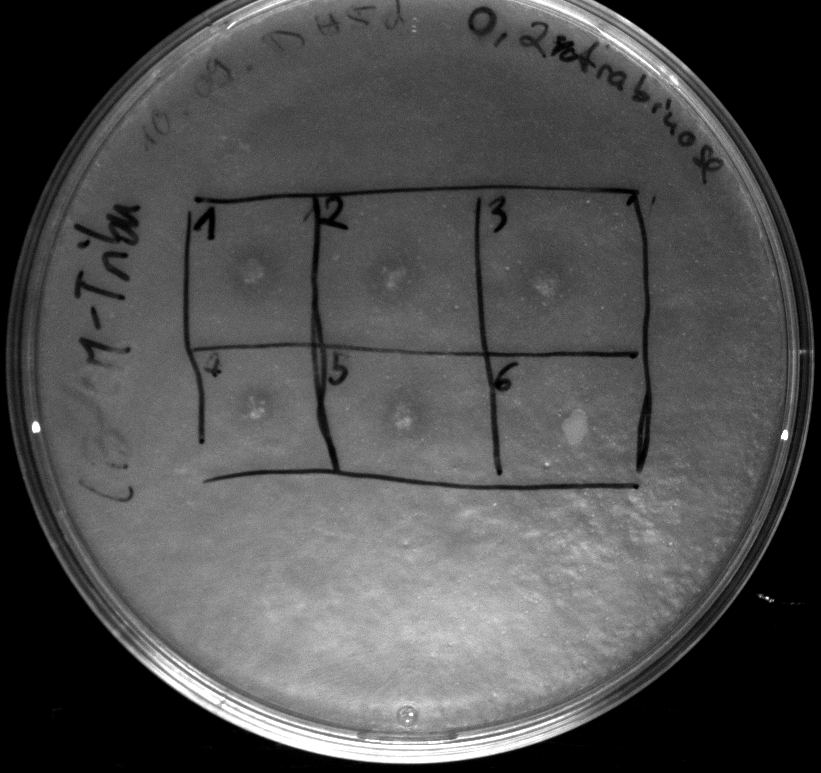
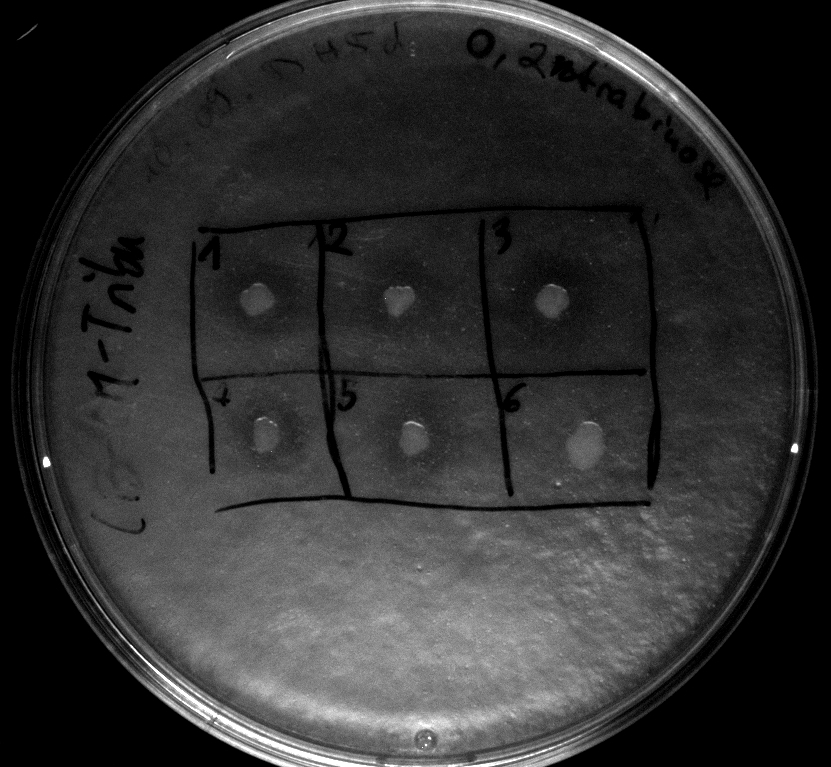
- activity assay of [http://ecoliwiki.net/colipedia/index.php/TOP10 Top10] containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808032 BBa_K808032] on LB Tributyrin agar with L-arabinose concentrations: 0.02%, 0.2%, 0.5%
- colonies 1-3 containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808032 BBa_K808032] but colony 4 containing [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808030 BBa_K808030]
- Protein purification from induced cultures and negative control
- cell debris pellets are treated with [http://www.sigmaaldrich.com/catalog/product/sigma/d4641?lang=en®ion=US n-Dodecyl ß-D-maltoside] as a detergent
- SDS PAGE (Laemmli) of pellets and supernatants
- Westernblot is successful as well
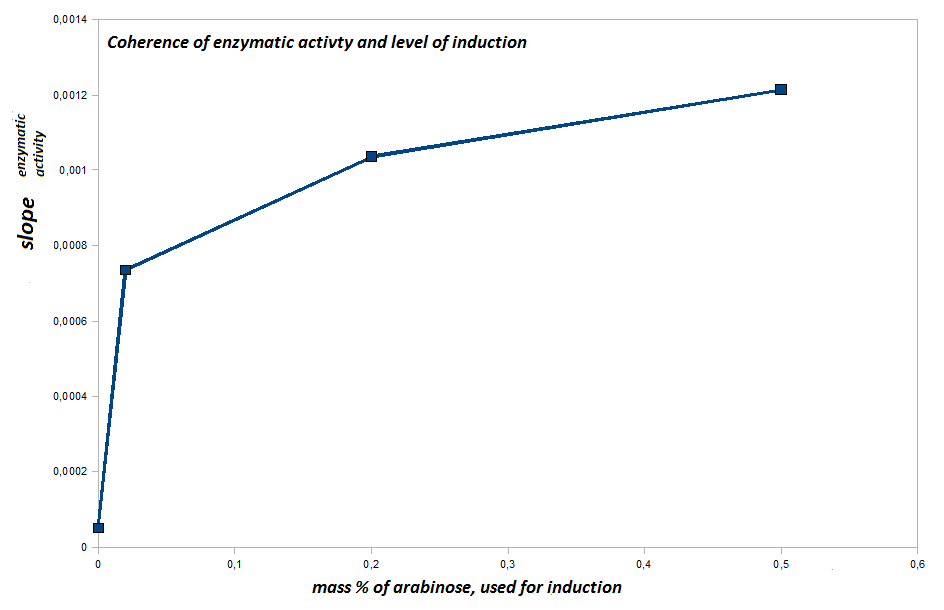
- to quantify the hydrolytic activity of our induced bacteria we perform a bacterial pNP-Assay.
We use our induced cells (Top10) with an OD600=0.1-0.025 (resuspended in PBS-buffer) as catalysts and para-Nitrophenylbutyrate as a substrate. Induced cells have the [http://partsregistry.org/Part:BBa_K808030 fusion protein (BBa_K808030)] expressed on their surface. The catalytical domain, formed by pNB-Esterase 13 will show hydrolytic activity we can screen for by performing a pNP-assay. The substrate will be hydrolysed, and one degradation construct is para-Nitrophenylbutyrate. If released by hydrolysis, a characeristic absorption at 405 nm can be detected and plotted against the time, resulting in straight lines. The hydrolytic activity is quantified by the slope of the respective straight line.

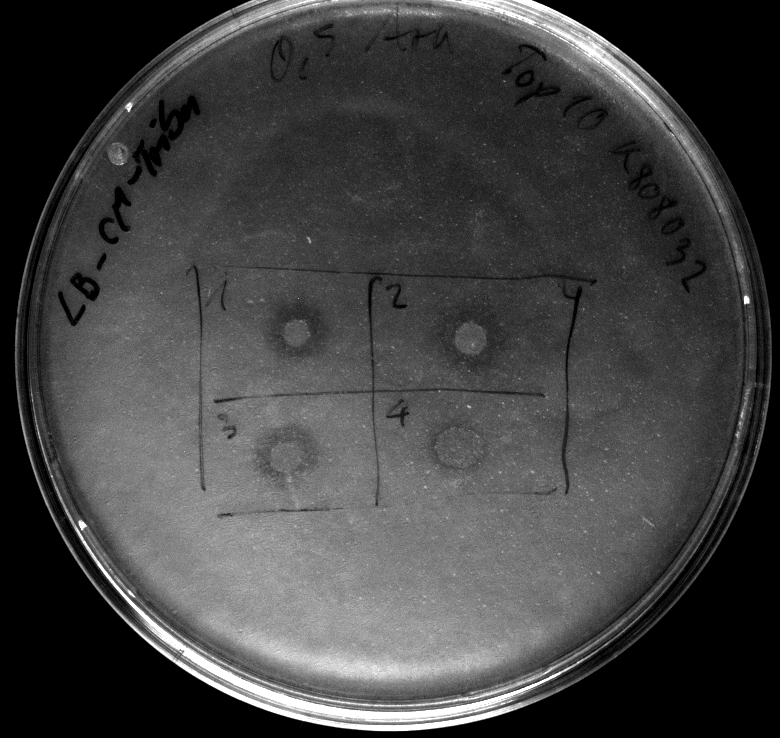
Wednesday, 19.09.12
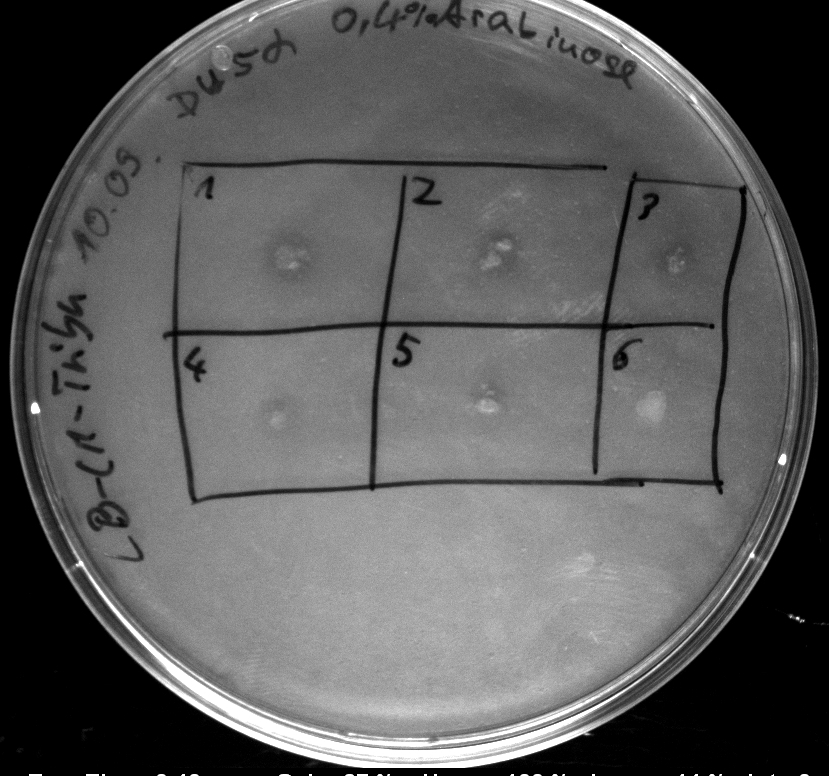
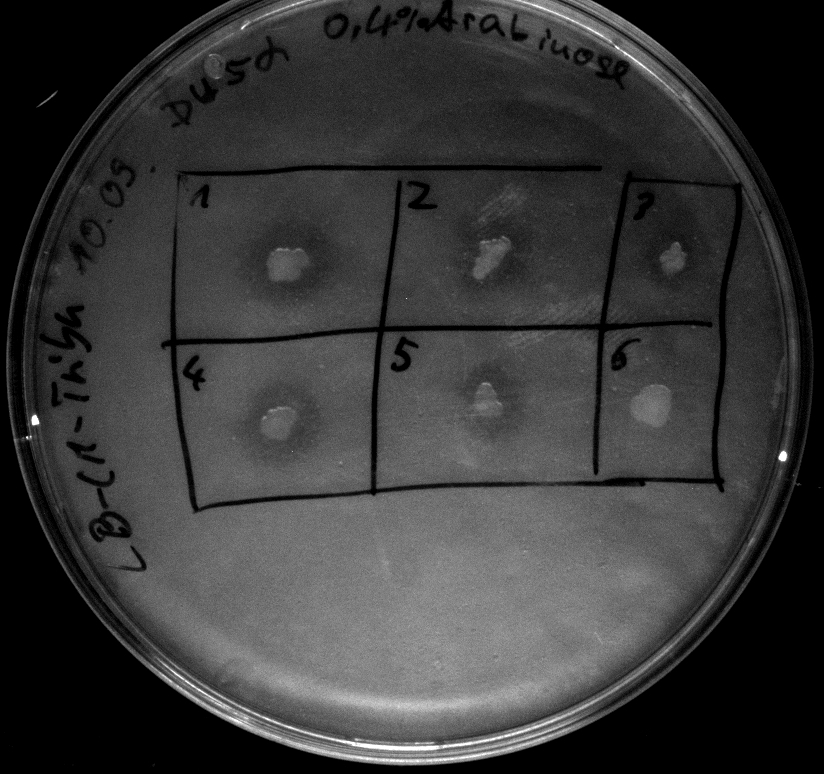
- activity assays on LB Tributyrin agar worked well (incubation occured just for the night at 30°C)
- activity assays on LB Tributyrin agar with 0.5% mw L-arabinose for inducing
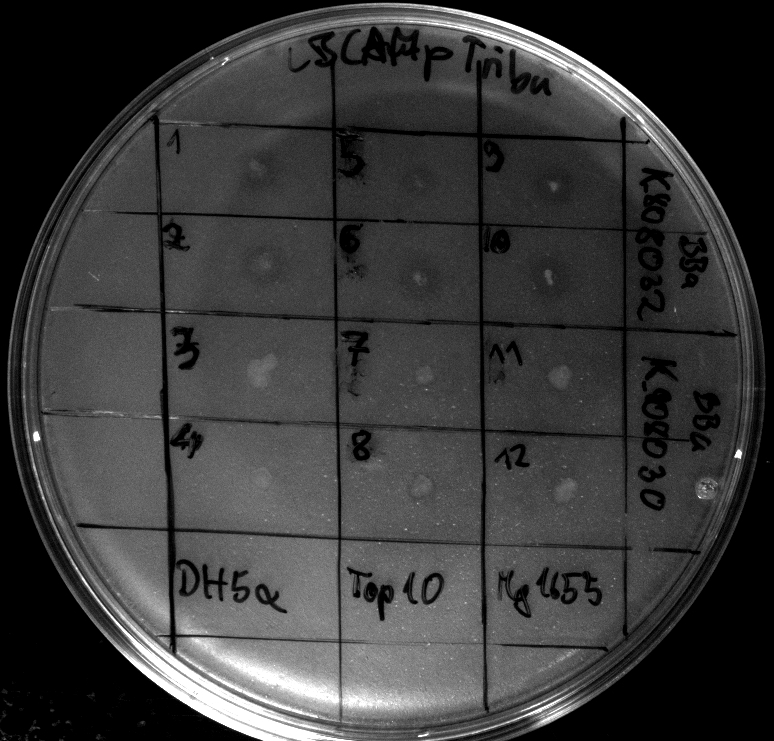
- colonies 1-4 [http://ecoliwiki.net/colipedia/index.php/DH5_alpha DH5 alpha], 5-8 [http://ecoliwiki.net/colipedia/index.php/TOP10 Top10], 9-12 [http://ecoliwiki.net/colipedia/index.php/MG1655 Mg1655]
- colonies 1,2,5,6,9,10 are carrying [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808032 BBa_K808032]
- colonies 3,4,7,8,11,12 are carrying [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808030 BBa_K808030]
Tuesday, 18.09.12
- activity assays on LB Tributyrin agar worked well
Protein Expression
CW 20
Monday 14.05.2012
- Bacterial transformation by heatshock of [http://www.neb.com/nebecomm/products/productc3029.asp BL21(DE3)SHuffle] with the plasmid pET26b(+)pNBDSM13 encoding Esterase 13 recieved by Henkel AG & Co. KGaA
- bacterial suspension was poured out on LB plates containing Cmp
Tuesday 15.06.2012
- one coclony of the retransformation was picked and transfered into 50 mL LB-Medium containing Cmp
- the culture was stirred at 180 rpm at 30°C
Wednesday 16.06.12
- centrifugation of the culture and resolving the pellet in 2 mL LB-Medium and 15% glycerine
- seperate the cell suspension into 100 µL aliquots
- store the stocks at -80°C
CW 24
Thursday, 14.06.2012
- PCR for protein expression of FsC
- each PCR is performed in 5 batches à 50 µL.
- Parameter: TA = 57°C, tA = 35 s, tE = 65 s
- PCR on pEST100 vector for expression with pEX vector gene of interest: FsC for designed part [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808032 BBa_K808032]
primers: pEX FsC His SfiI up and pEX FsC Stop SfiI lo
Friday, 15.06.2012
- Agarose gel electrophoresis for quality control
CW 25
Wednesday, 20.06.2012
- Agarose gel electrophoresis for preparation
- Gel extraction of PCR products
- Concentration of produced FsC sequence: 40 ng/µL
Thursday, 21.06.2012
- Restriction digest of PCR product from 14.06.
- Enzymes used: SfiI
- NEBuffer: 4
- Digestion time: over night
- Digestion temperature: 50°C
Friday, 22.06.2012
- Miniprep of pEX vector
- Concentration range: 240-488 ng/µL
- Restriction digest of pEX
- Enzymes used: SfiI
- NEBuffer: 4
- Digestion time: 3 days
- Digestion temperature: 50°C
CW 26
Monday, 25.06.2012
- Agarose gel electrophoresis for quality control
- Gel extraction of pEX and digested FsC sequence from 21.06.
- Concentrations: pEX: 77 ng/µL, FsC sequence: 18 ng/µL
- Ligation of pEX with FsC sequence with the ratio 1:3 and 1:5
| Component | 1:3 | 1:5 |
|---|---|---|
| FsC sequence | 1.12 µL | 2.2 µL |
| pEX | 0.64 µL | 0.64 µL |
| Ligase buffer | 4 µL | 4 µL |
| T4 DNA ligase | 1 µL | 1 µL |
| H2O | 33 µL | 30 µL |
- Ligation time: over night
Tuesday, 26.06.2012
- Bacterial transformation by Electroporation of TOP10 with the ligation product pEX-FsC from 25.06.
Wednesday, 27.06.2012
Thursday, 28.06.2012
- Miniprep of pEX-Fsc in TOP10
- Concentration: 45 ng/µL and 405 ng/µL
- Bacterial transformation by Electroporation of [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/bacterial-strains-bmh-71_18-muts-and-es1301-muts/ BmH7118] with the Miniprep product pEX-FsC
CW 27
Monday, 02.07.2012
- Protein expression of FsC at different temperatures (16°C, 25°C, 30°C and 37°C) for the final step of the expression
Tuesday, 03.07.2012
- Purification of Periplasmatic Proteins for all expression temperatures
Wednesday, 04.07.2012
- Protein purification without cell disrupter
- SDS-PAGE (Schägger) analysis of all collected samples
| Expression at 16°C/25°C |
|---|
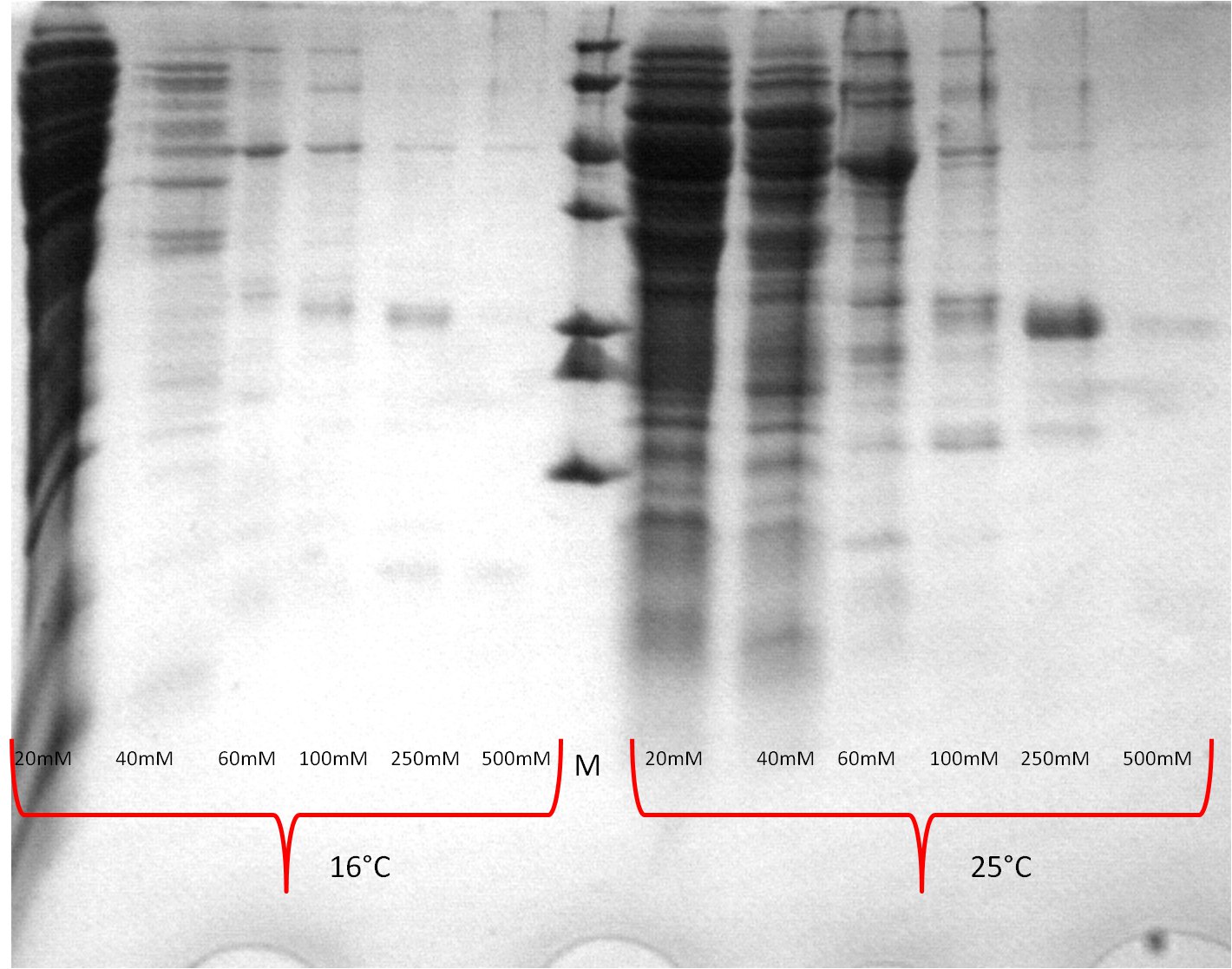
|
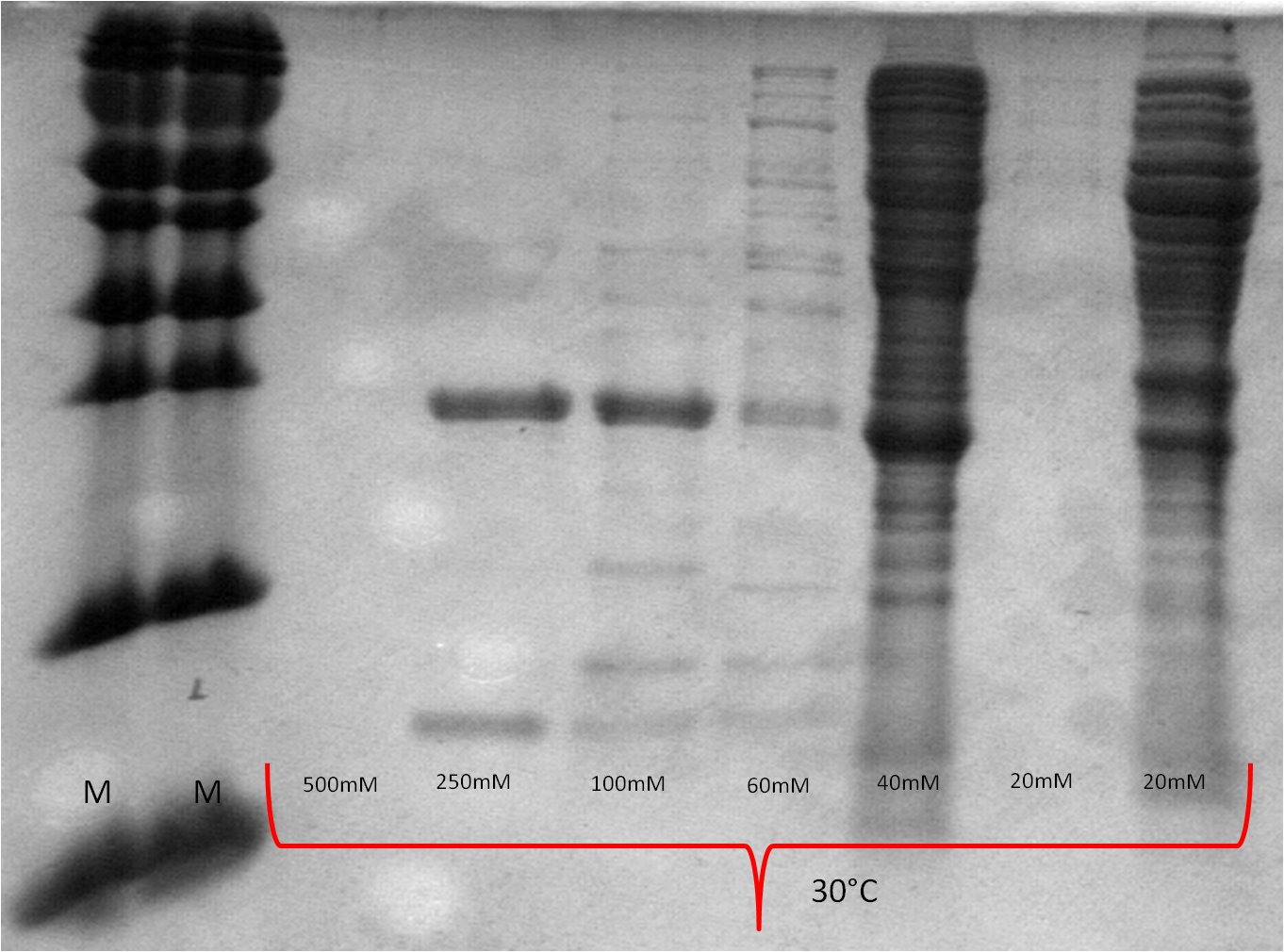
| Expression at 30°C/37°C |
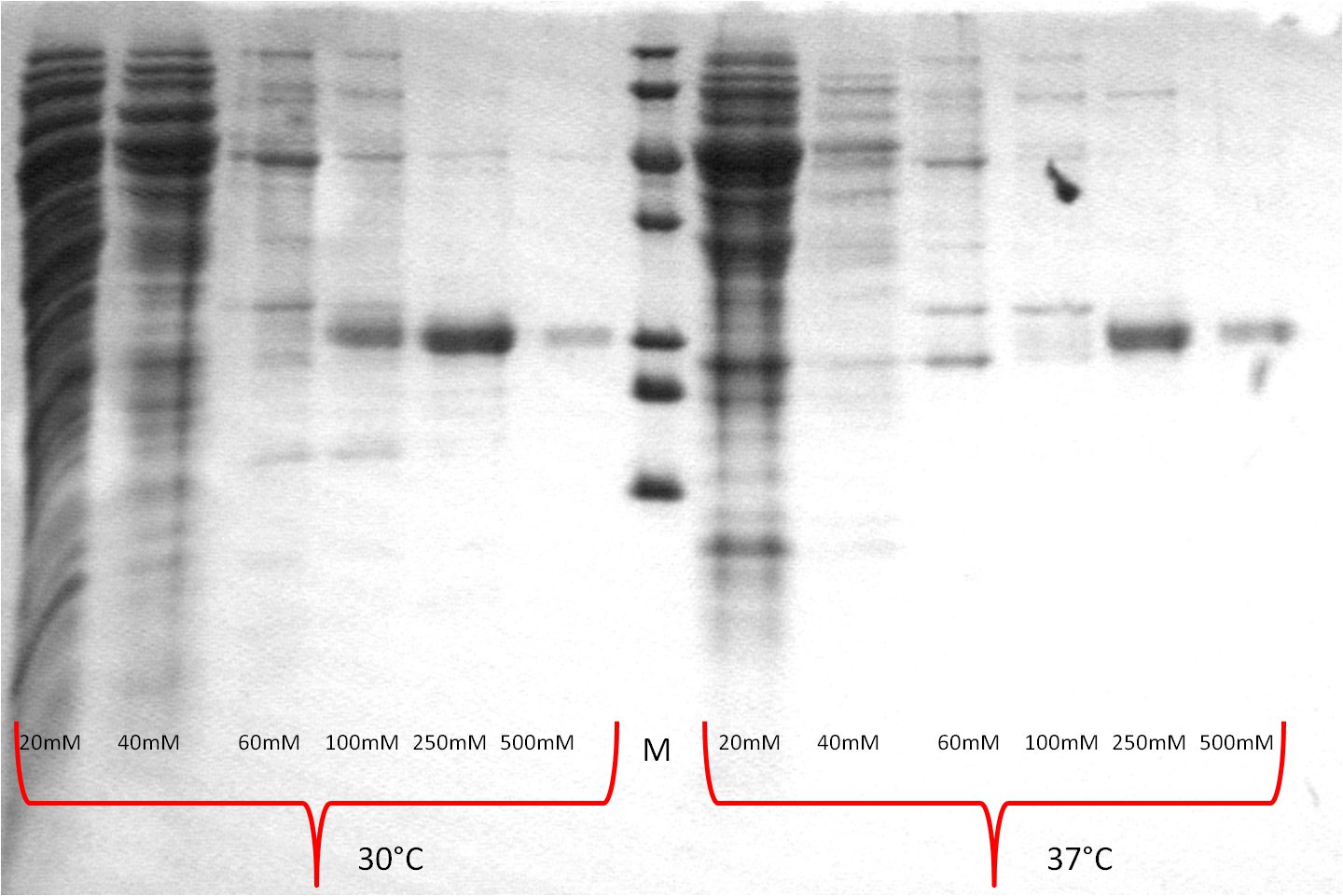
|
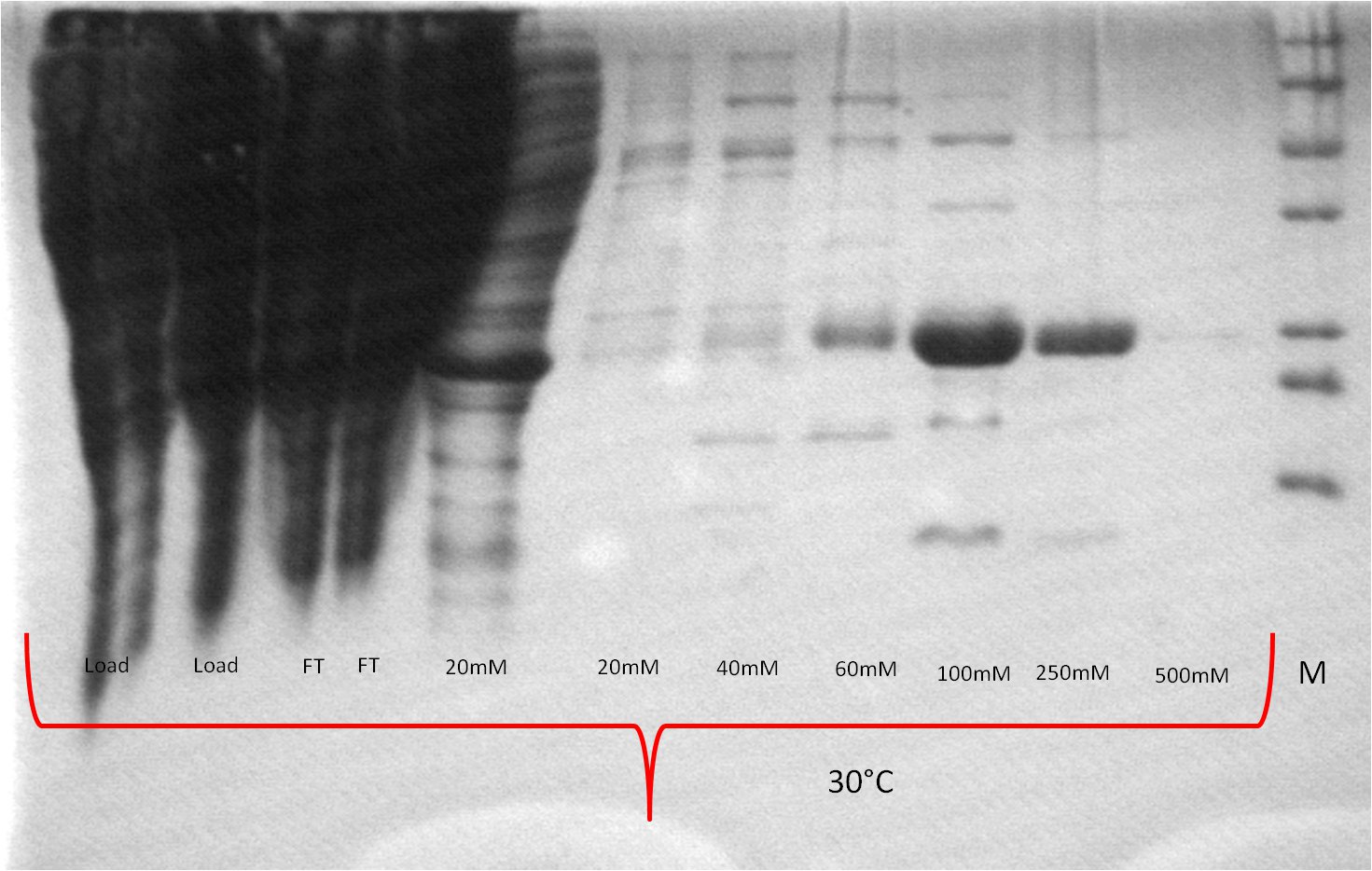
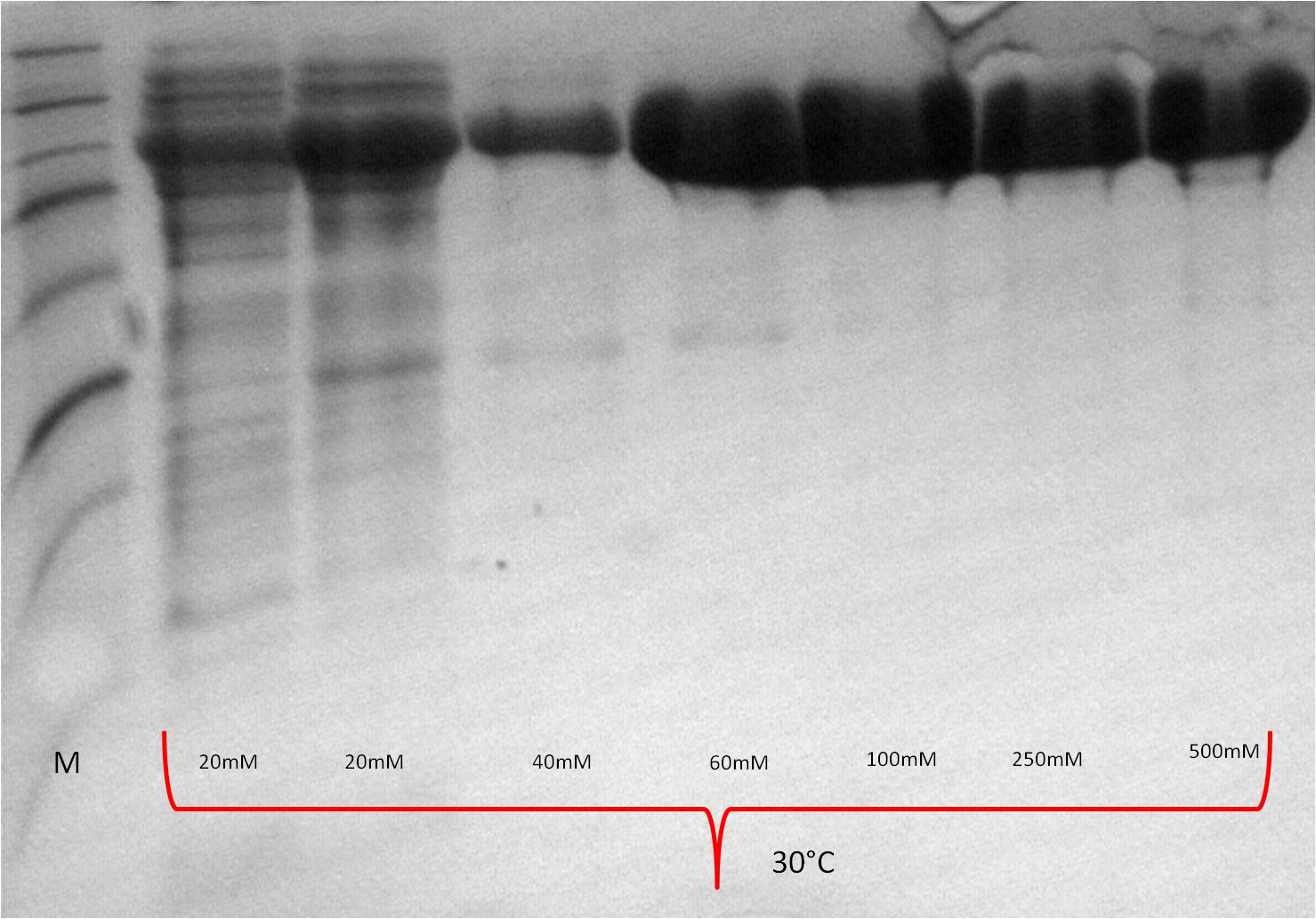
- The best results were maintained at an expression temperature of 30°C
Friday, 06.07.2012
- Inoculation of 5 ml DYT Cmp with [http://www.promega.com/products/cloning-and-dna-markers/cloning-tools-and-competent-cells/bacterial-strains-and-competent-cells/bacterial-strains-bmh-71_18-muts-and-es1301-muts/ BmH7118] containing pEX-FsC
- Incubation for 3 days at 20°C
CW 28
Monday, 09.07.2012
- Miniprep of pEX-FsC from 06.07.
- Concentration: 470 ng/µL
- Preparation for sequencing at Eurofins
- Barcode 043 pEX-FsC with primer M13 Reverse up
- Barcode 044 pEX-FsC with primer ClaI pIII lo
- Plasmid DNA: 5µL
- Primer: 3µL
- H2O: 7µL
Wednesday, 11.07.2012
- Protein expression of FsC according to standard protocol
Thursday, 12.07.2012
- Protein purification by standard protocol
- SDS-PAGE (Schägger) analysis of samples
CW 29
Monday, 16.07.2012
- Protein expression of Est13 according to standard protocol
Tuesday, 17.07.2012
- Protein purification by standard protocol
- SDS-PAGE (Schägger) analysis of samples
CW 31
Monday, 30.07.2012
- Protein expression of FsC according to standard protocol
Tuesday, 31.07.2012
- Protein purification by standard protocol
- SDS-PAGE (Schägger) analysis of samples
CW 33
Friday, 17.08.2012
- Protein expression of FsC according to standard protocol
Saturday, 18.08.2012
- Protein purification by standard protocol
- SDS-PAGE (Schägger) analysis of samples
CW 34
Wednesday, 22.08.2012
- Protein expression of FsC according to standard protocol
Thursday, 23.08.2012
- Protein purification by standard protocol
- SDS-PAGE (Schägger) analysis of samples
Enzyme Activity Assays
To characterise our BioBricks [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808026 BBa_K808026] (pNB-Est13) & [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808025 BBa_K808025] (FsC) a pNP-assays are performed.
CW 37
Est13
- a pNP-assayis performed with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808026 BBa_K808026] (pNB-Est13)
Km and Vmax calculation
To find Km the growth of the first seven data points were written down in a diagram. The slope of the regression line gave the speed of the hydrolysis for the single substrate concentrations. Now the value of the speed with the associated concentration of substrate were plotted in a Lineweaver-Burk diagram. At the point where the regression line is crossing the x-axis the x-coordinate amounts to -1/Km. So Km is about 400µM. To calculate the value of Vmax we used the point where the plotted line crosses the y-axis. The associated y-coordinate is 1/Vmax. Consequently Vmax for 50nM Est13 is 0,002798 1/s.
Kcat calculation
The value Kcat is calculated through dividing Vmax by the enzyme concentration of 50nM. Kcat counts 0,055951 1/mol*s.
FsC
- a pNP-Assay is performed with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K808025 BBa_K808025].
Km and Vmax calculation
To find Km the growth of the first twelve data points were written down in another diagram. The slope of the regression line gave the speed of the hydrolysis for the single substrate concentrations. Now the value of the speed with the associated concentration of substrate were plotted in a Lineweaver-Burk diagram. At the point where the regression line is crossing the x-axis the x-coordinate amounts to -1/Km. So Km is about 400µM. To calculate the value of Vmax we used the point where the plotted line crosses the y-axis. The associated y-coordinate is 1/Vmax. Consequently Vmax for 5nM FsC is 0,001467 1/s.
Kcat calculation
The Kcat value is calculated through dividing Vmax by the enzyme concentration of 5nM. Kcat counts 0,293395 1/mol*s.
FsC with ehtylenglycol
Our Simulation lab was able to show , theoratically, a relation between an decrease of enzyme activity and an increase of ethylenglycol concentration. This could be a hint for the mysterious loss of hydrolysis when the degradation rate reaches 5% of PET foil weight as it is described in the literature (Ronkvist, Å. M., Xie, W., Lu, W., & Gross, R. a. (2009). Cutinase-Catalyzed Hydrolysis of Poly(ethylene terephthalate). Macromolecules, 42(14), 5128–5138. doi:10.1021/ma9005318). We tried to proof their theory of potentially higher ethylenglycol concentration surrounding the PET foil. An increasing amount of ehtylenglycol should hamper the hydrolytic activity of FsC in a pNP-assay.
Procedure
The lab test procedure was nearly equal to the normal kinetics records with only one difference. The reaction solution consisted of different composites of PBS buffer and ethylenglycol (0 - 20%). All records were done at a concentration of 1mM of 4-Nitrophenyl butyrate in the experiment.
Records of the different ethylenglycol concentrations and the associated speeds of hydrolisis in a diagram
Interpretation
As you can see there is a stagnation of the speed of hydrolysis of the ester caused by an increasing concentration of ethylenglycol. But by that you cannot tell that ethylenglycol inhibits the FsC, because the hydrolysis of the ester rose with a higher concentration of ethylenglycol in the negative samples. So it could be that the ethylenglycol starts a concurrent reaction or blocks the 4-Nitrophenyl butyrate.
CW 36 - 38
PET degradation
Principle and conditions

Conditions: Room temperature (~ 22°C) pH = 7.4
Procedure
Round about 1g of PET granulate is added in each two falcon tubes to between 15 and 20 mL water of pH = 7.4 and several drops of [http://en.wikipedia.org/wiki/Bromothymol_blue bromothymol blue]. In one of these falcon tubes Est13 stock solution is added until a concentration of 2,5 µM is reached. For 15 days the solutions are incubated at room temperature. At nine specific days of these the enzymatic solutions and the negative sample are photographed, to control the shift of pH.
Results
In all photos the degradation with Est13 is in the left falcon tube.
The photos show a decrease of the pH-value over time in the sample containing Est13. The one without this enzyme remains unchanged. So it is concluded that Est13 degrades PET granulate.
CW34 - CW35
Degradation of monomethyl terephthalate over time
Principle and conditions
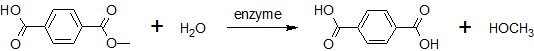
Conditions: Room temperature (~22°C)
Procedure
Three different samples of 30 mL dest. water at a pH of 7.4 are incubated for 7 days with a concentration 500µM of monomethyl terephthalate (solved in methanol) and following incredents:
1. 30µL of FsC (concentration in the incubated solution: ~0,8µM)
2. 30µL of Est13 (concentration in the incubated solution: ~0,8µM)
3. Nothing
To control the shift of the pH during the estercleavage some drops of [http://en.wikipedia.org/wiki/Bromothymol_blue bromothymol blue] are added.
Results
Following pictures show the shift of the pH over 7 days (The degaradation with Est13 is in the left falcon tube and with FSC in the middle one):
So you can see that both enzymes catalysed the cleavage of the ester in a few days. In 7 days the pH shifted about one unit.
 "
"