Team:HokkaidoU Japan/Notebook/plastic Week 9
From 2012.igem.org
July 2nd
Digestion
Digestion to divide BBa_K342001(PhaC) with XbaI and PstI.
And BBa_B0034(RBS) with SpeI and PstI (with three samples).
PhaC (BBa_K342001)
| DNA solution (100 ng/ul) | 12.5 ul |
| XbaI | 1 ul |
| PstI | 1 ul |
| 10xM buffer | 2 ul |
| DW | 3.5 ul |
| Total | 20 ul |
RBS (BBa_B0034)
N0.1
| DNA solution (20.3 ng/ul) | 14.3 ul |
| SpeI | 1 ul |
| PstI | 1 ul |
| 10xH buffer | 2.5 ul |
| DW | 0.2 ul |
| Total | 25 ul |
N0.2
| DNA solution (15.6 ng/ul) | 18.6 ul |
| SpeI | 1 ul |
| PstI | 1 ul |
| 10xH buffer | 2.5 ul |
| DW | 1.9 ul |
| Total | 25 ul |
N0.3
| DNA solution (16.9 ng/ul) | 17.2 ul |
| SpeI | 1 ul |
| PstI | 1 ul |
| 10xH buffer | 2.5 ul |
| DW | 3.3 ul |
| Total | 25 ul |
Gel extraction
Gel extraction for digestion product. Used FastGene Gel&PCR extraction kit(NipponGenetics)and got 50 ul of DNA solution.
August 22th
Mini-prep
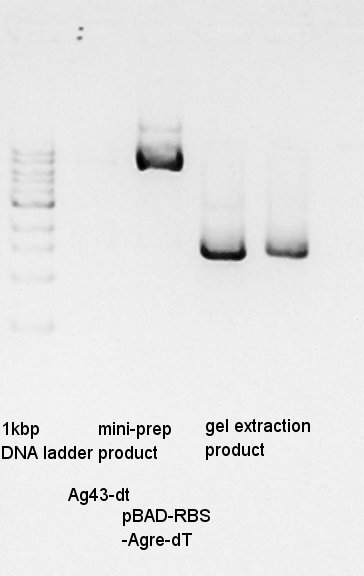
Mni-prep of pT7-RBS on pSB1C3 of colony No.1 and 2 selected by the result of colony PCR in 20th. We used mini-prep kit of Nippon genetics: FastGene Plasmid mini kit and finally we eluted the DNA with 20 ul of elution buffer.
One of Ag43-dT on pSB1AK3 culture did not get myddy. And another one is only a little muddy. We tried mini-prep to the latter, we god the 20 ul of DNA solution. And then, we did electrophoresis the mini-prep products and (pBAD-RBS and pBAD) gel extract products.
PCR
PCR of pT7-RBS on pSB1C3.
We used 4 kinds of primer set.
1 : EX-F , PS-R primer
2 : EX-F , 200b down primer
3 : 100b up , PS-R primer
4 : 100b up , 200b down primer
The density of primer solutions is 10 uM.
| DNA solution | 1 ul |
| KOD-Plus-NEO(Taq polymerase) | 1 ul |
| dNTP | 5 ul |
| MgSO4 | 3 ul |
| KOD-Plus-NEO Buffer | 5 ul |
| Forward Primer | 1 ul |
| Reverse Primer | 1 ul |
| DW | 33 ul |
| Total | 50 ul |
| Number | Degree | Second |
| 1 | 94 | 120 |
| 2 | 98 | 10 |
| 3 | 58 | 30 |
| 4 | 68 | 30 |
| 5 | 4 | HOLD |
Cycle:2~4 x 45
[[image:|thumb|PCR result]]
August 23th
Ethanol precipitation
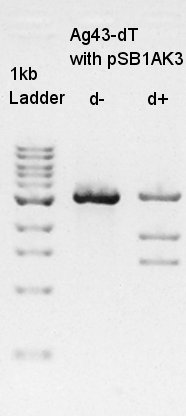
Ethanol precipitation to get more high concentration of Ag43-dT on pSB1AK3 solution cut with XbaI & SpeI.
- Added 2 ul of NaoAc, 1.5 ul of glycogen and 50 ul of 100% ethanol.
- Centrifuged in 15000 rpm, 10 min at 4C.
- Remove supernatant and added 220 ul of 70% ethanol.
- Centrifuged in 15000 rpm, 10 min at 4C.
- Remove supernatant and air drying in room temperature then added 10 ul of DW.
Digestion
Digestion to divide Ag43-dT and pSB1AK3 which has same number of bp as Ag43-dT by cut with HindIII.
| DNA solution ( 257ng/ul) | 9 ul |
| HindIII(15U/ul) | 1 ul |
| 10xM buffer | 2 ul |
| DW | 8 ul |
| Total | 20 ul |
| Number | Degree | Minute |
| 1 | 37 | 180 |
| 2 | 70 | 15 |
| 3 | 4 | HOLD |
In this result, we confirmed that the pSB1AK3 was successfully digested with HindIII, but it was not clear how many pSB1AK3 were remaining as non-digested products.
Gel extraction
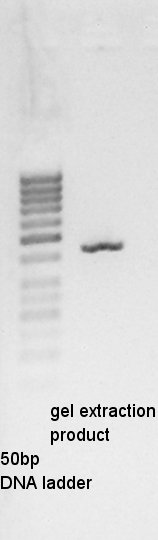
Gel extraction for digestion product. We used FastGene Gel&PCR extraction kit(NipponGenetics)and got 50 ul of DNA solution.
August 24th
Digestion
Digestion of pT7-RBS on pSB1C3 with SpeI, Ag43-dT on pSB1AK3 with EcoRI & XbaI and pBAD-RBS with EcoRI & PstI. Ag43-dT on pSB1AK3 E&X
| DNA solution ( 120ng/ul) | 7 ul |
| EcoRI | 1 ul |
| XbaI | 1 |
| 10xM buffer | 2 ul |
| DW | 9 ul |
| Total | 20 ul |
E (control)
| DNA solution ( 120ng/ul) | 7 ul |
| EcoRI | 1 ul |
| 10xM buffer | 2 ul |
| DW | 10 ul |
| Total | 20 ul |
X (control)
| DNA solution ( 120ng/ul) | 7 ul |
| XbaI | 1 |
| 10xM buffer | 2 ul |
| DW | 10 ul |
| Total | 20 ul |
pBAD-RBS(E & S)
| DNA solution ( 100ng/ul) | 12 ul |
| EcoRI | 1 ul |
| SpeI | 1 |
| 10xH buffer | 2 ul |
| DW | 4 ul |
| Total | 20 ul |
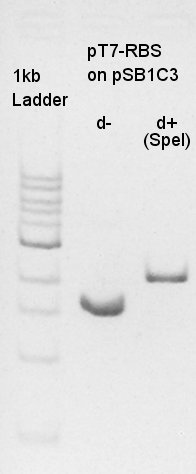
pT7-RBS on pSB1C3 (SpeI)
| DNA solution ( 20ng/ul) | 4 ul |
| SpeI | 1 ul |
| 10xM buffer | 2 ul |
| DW | 13 ul |
| Total | 20 ul |
| Number | Degree | Minute |
| 1 | 37 | 180 |
| 2 | 60 | 15 |
| 3 | 4 | HOLD |
About pT7-RBS on pSB1C3, we successfully digested the plasmid DNA and converted it to linear DNA.
Gel extraction
Gel extraction for digestion product. We used FastGene Gel&PCR extraction kit(NipponGenetics)and got 50 ul of DNA solution.
Ethanol Precipitation
Ethanol precipitation for pT7-RBS on pSB1C3 and Ag43-dT digestion products.
- Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol.
- Centrifuged in 15000 rpm, 10 min at 4C.
- Remove supernatant and added 220 ul of 70% ethanol.
- Centrifuged in 15000 rpm, 10 min at 4C.
- Remove supernatant and air drying in room temperature then added 5 ul of DW.
Ligation
Ligation for Ag43-dT as insert and pT7-RBS on pSB1C3 as vector. We use Ligation Mighty Mix (TAKARA BIO INC.) which contains ligase and buffer.
| Vector DNA | 4 ul |
| Insert DNA | 4 ul |
| DW | 2 ul |
| Ligation Mighty Mix | 10 ul |
| Total | 20 ul |
Ligation reaction time was in detail below.
| Degree | Minute |
| 16 | 30 |
| 65 | 10 |
| 4 | Hold |
Transformation
Transformation for ligation product. We used E.coli strain DH5α.
- Added 2 ul of DNA to 50 ul of thawed competent cells on ice.
- Incubated on ice for 30 min.
- Added 350 ul of LB.
- Incubated the cells for 2 hours at 37C.
- Prepared and Labeled two plastic plates with LBC.
- Plated 300 ul of the culture onto first dish and spread.
- Added 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread.
- Incubated the plates at 37C for hours.
August 25th
Colony PCR
Colony PCR to confirm that whether the pT7-RBS-Ag43-dT on pSB1C3 was successfully ligated or not.
| DNA solution | 4 ul |
| Kapa-Taq(Taq polymerase) | 5 ul |
| Forward Primer(ag43-f4 primer) | 0.5 ul |
| Reverse Primer(200bp down primer) | 0.5 ul |
| Total | 10 ul |
| Number | Degree | Second |
| 1 | 95 | 120 |
| 2 | 95 | 30 |
| 3 | 53.0 | 30 |
| 4 | 72 | 60 |
| 5 | 72 | 60 |
| 6 | 4 | HOLD |
Cycle:2~4 x 35
We used N1 (DW only) and N2 (Ag43-dT on pSB1AK3) as controls. Desired product is about 695bp.
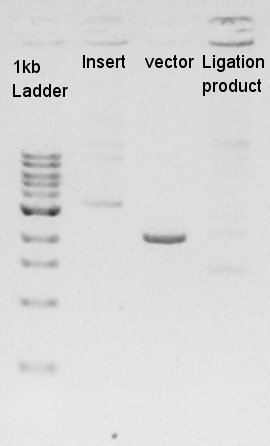
The results showed that desired DNA were not existed in these picked up colonies. We observed our ethanol precipitation and ligation products electrophoresis result image (see August 24th) then noticed that the concentration of each ethanol precipitation products were reversed. This means vector DNA would be self-ligated by the high ratio of molar in ligation reaction solution. We decided to try the DNA synthesis from digestion reaction of vector DNA once more time.
Digestion
Digestion of vector DNA: pT7-RBS on pSB1C3 with SpeI. Not to leave the plasmid DNA as plasmid DNA, we cut the DNA in overnight. pT7-RBS on pSB1C3 (SpeI)
| DNA solution ( 20ng/ul) | 4 ul |
| SpeI | 1 ul |
| 10xM buffer | 2 ul |
| DW | 13 ul |
| Total | 20 ul |
| Number | Degree | Minute |
| 1 | 37 | 600 (10 hours) |
| 2 | 60 | 15 |
| 3 | 4 | HOLD |
August 26th
Gel extraction
Gel extraction for digestion product. We used FastGene Gel&PCR extraction kit(NipponGenetics)and got 50 ul of DNA solution.
Ethanol Precipitation
Ethanol precipitation for pT7-RBS on pSB1C3 and Ag43-dT digestion products.
- Added 5 ul of NaoAc, 1.5 ul of glycogen and 125 ul of 100% ethanol.
- Centrifuged in 14000 rpm, 30 min at 4C.
- Remove supernatant and added 220 ul of 70% ethanol.
- Centrifuged in 15000 rpm, 15 min at 4C.
- Remove supernatant and air drying in room temperature then added 5 ul of DW.
Ligation
Ligation for Ag43-dT as insert and pT7-RBS on pSB1C3 as vector. We use Ligation Mighty Mix (TAKARA BIO INC.) which contains ligase and buffer.
| Vector DNA | 0.25 ul |
| Insert DNA | 3 ul |
| DW | 1.75 ul |
| Ligation Mighty Mix | 5 ul |
| Total | 20 ul |
Ligation reaction time was in detail below.
| Degree | Minute |
| 16 | 30 |
| 65 | 10 |
| 4 | Hold |
Transformation
Transformation for ligation product. We used E.coli strain DH5α.
- Added 2 ul of DNA to 50 ul of thawed competent cells on ice.
- Incubated on ice for 30 min.
- Added 350 ul of LB.
- Incubated the cells for 2 hours at 37C.
- Prepared and Labeled two plastic plates with LBC.
- Plated 300 ul of the culture onto first dish and spread.
- Added 450 ul of LB to 50 ul of the culture and plated 300 ul of it onto second dish and spread.
- Incubated the plates at 37C for hours.
 "
"