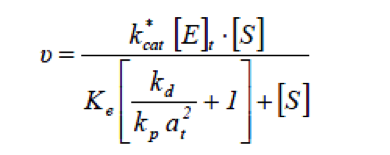Team:NTU-Taida/Modeling/FA
From 2012.igem.org
Fatty Acid Reaction Absorption Model
Contents |
Overview
With the single cell model, we are able to simulate the response time of our system after the concentration of fatty acid exceeds our filtering threshold. However, the overall response time is also determined by the time needed for the eaten fat to be hydrolysis and producing fatty acid with concentration higher than the threshold.
Furthermore, since our E.coli cells mainly reside on the intestinal walls instead of the lumen, we have to determine the actual concentration of fatty acid around the intestinal wall, which is different from that in the lumen space.
Therefore we performed a simulation to model the hydrolysis of fat by lipase into glycerol and fatty acids. In parallel, we investigated the mechanism of long chain fatty acid absorption and constructed a two dimensional spatial-temporal model with COMSOL Multiphysics. Finally, we incorporate the reaction of fat hydrolysis into our spatial-temporal model describing fatty acid absorption and therefore result in a combined model simulating the reaction and absorption of fatty acid around the intestinal wall.
Reaction Model
Background
We start our effort for simulating the hydrolysis of fat by determining the rate of fat hydrolysis catalyzed by lipase.
Lipase catalysed reaction take place at the interface between the aqueous phase containing the enzyme and the oil phase, therefore the rate of hydrolysis by lipase will depend on the total specific interfacial area.[1] When considering systems in which the total specific interfacial area changes with changes in operating condition, the rate of fat hydrolysis can be given by
- at
- Total specific interfacial area [m-1]
Equations
Results
Combined Reaction-absorption Model
Spatial-temporal Model Equations
Model Design
Results
Simulation for longer time span
Reference
 "
"