Team:Bielefeld-Germany/Labjournal/week21
From 2012.igem.org
Contents |
Week 21 (09/17 - 09/23/12)
Monday September 17th
- Team Cultivation & Purification:
- Cell disruption of fermentation of E. coli Rosetta-Gami 2 with [http://partsregistry.org/wiki/index.php?title=Part:BBa_K863012 BBa_K863012] via high-pressure homogenization and purification via Ni-NTA column.
Tuesday September 18th
- Team Cellulose Binding Domain:
- Primer arrived:
- Gradient-PCR with the [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His]-plasmid and S3N10-CBDcex_RV and S3N10-GFP_FW primer-mix. the right product appeared only at 66,5°C; Lower: wrong product; Higher: no product
- Following PCR at 66,5°C(50µL) had the wrong product (main product at 2 kbp); could be because of a different template concentration or not exact temperature.
- Alternativly took the 20 µL-tube with right product and added 0,5 µL DpnI for over-night digestion.
- Gradient-PCR with [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His]-template and a CBDcex_Freiburg- + GFP_Freiburg_compl-primer-mix to amplify the whole fusion-protein and get rid of the His-tag.
- Gradient-PCR with [http://partsregistry.org/Part:BBa_K863113 CBDclos(T7)+GFP_His]-template and a CBDclos_Freiburg- + GFP_Freiburg_compl-primer-mix to amplify the whole fusion-protein and get rid of the His-tag.
- Team Substrate Anayltic: We analyze Ethin estradiol and estradiol degradation with the Spectrofluorophotometer. The measure showed that the degradation product are detectable but the results weren't clear. We decide to do this measurement again to have better results so a new degradation of the two substrates were set.
Wednesday September 19th
- Team Cellulose Binding Domain:
- Stopped over-night-DpnI-digestion of S3N10-clos-GFP_his PCR-product and cleaned up in gel. Ligated the PCR-product with itself via Blunt-end-ligation and transformed it into KRX. Plated on Select-Agar.
- Gradient-PCRs of the [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His]-plasmid and a [http://partsregistry.org/Part:BBa_K863113 CBDclos(T7)+GFP_His]-plasmid (unsequenced) with the CBDcex_Freiburg- and GFP_Freiburg-compl- and the CBDclos_Freiburg- and GFP_Freiburg_compl primers, respectively. The CBDcex-setup showed no correct product, but the CBDclos-setup had the right bands at temperatures from 57°C to 64°C; Merged the correct fractions and cleaned them up through the gel.
- Digested the cleaned-up product with SpeI and XbaI and DpnI.
- Ligated <partinfo>J61101</partinfo> and [http://partsregistry.org/Part:BBa_K863111 CBDclos_Freiburg] with [http://partsregistry.org/Part:BBa_K863120 GFP_Freiburg] and transformed it into KRX.
- Team Activity Test:
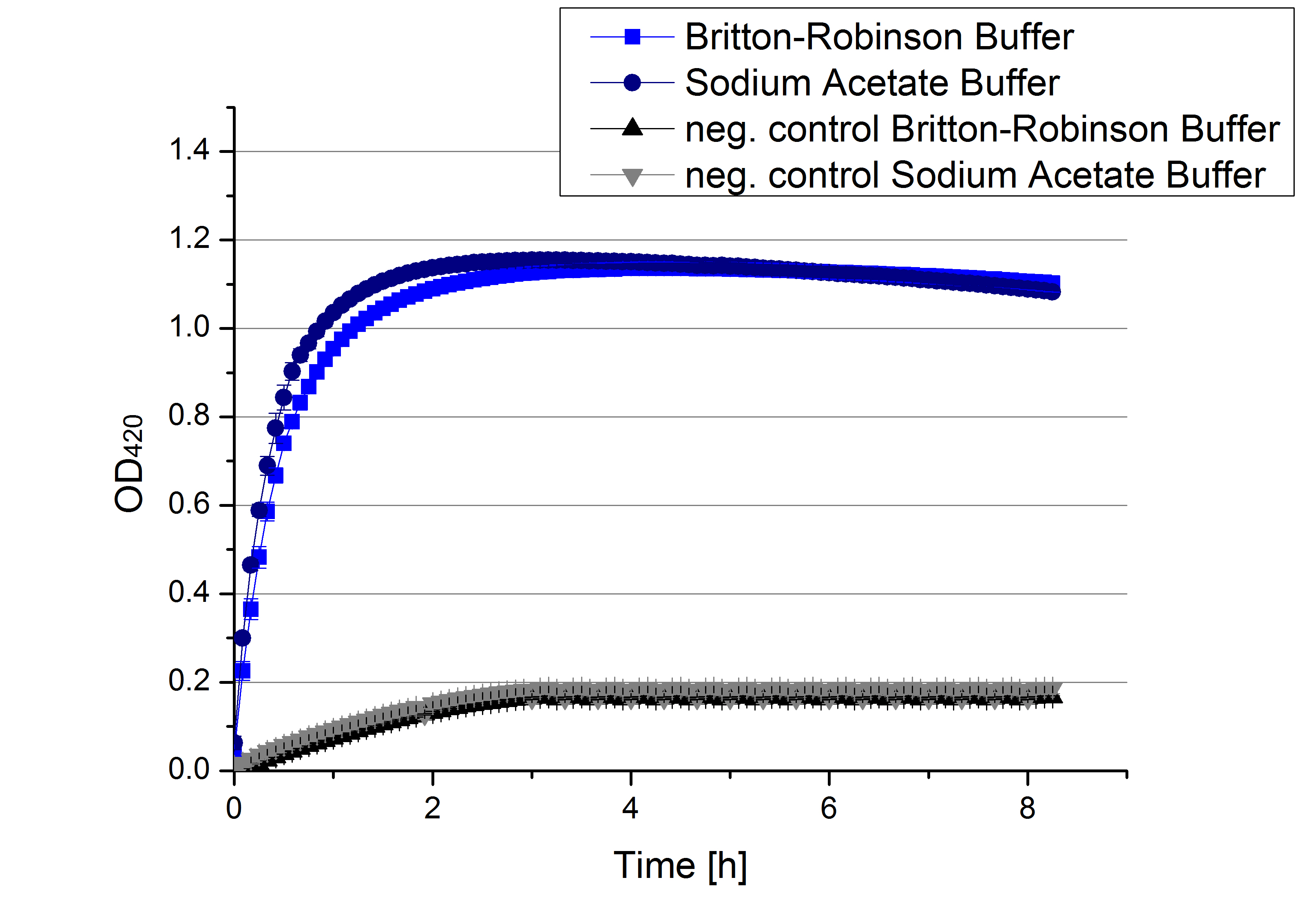
- What a day! The deadline is coming closer and we still had (!) a lot of measurements to do, so today was chosen to be a day full of important measurements. First we measured the activity of the TVEL0 and [http://partsregistry.org/wiki/index.php/Part:BBa_K863000 BPUL] in a CuCl gradient. As well we examined what our [http://partsregistry.org/wiki/index.php/Part:BBa_K863000 BPUL] laccase does when its supposed to work in different pHs. [http://partsregistry.org/wiki/index.php/Part:BBa_K863000 BPUL] was additionally characterized regarding its behavior in different concentrations of MeOH and acetonitrile. This output is of course most interesting for Team Substrate Analytics. Since we already compared the behaviour of TVEL0 in the environment of sodium acetate buffer (pH 5) and Britton-Robinson buffer (pH 5), respectively, we also compared [http://partsregistry.org/wiki/index.php/Part:BBa_K863000 BPUL] and [http://partsregistry.org/Part:BBa_K863005 ECOL] in those buffers. ECOL is not impressed by the two different buffer systems and thus not influenced in it´s activity.
Thursday September 20th
- Team Cellulose Binding Domain:
- No Colonies on the S3N10-[http://partsregistry.org/Part:BBa_K863113 CBDclos(T7)+GFP_His]-transformations-dish with the blunt-end ligation; reason: forgotten to phosphorylate the PCR-product.
- Two colonies on the CBDclos+GFP-dish with the constitutive [http://partsregistry.org/Part:BBa_J61101 J23100 + J61101] promoter. But none of them is glowing. And them shouldn't ... used the wrong selection-agar (since J61101 carries pSB1A2-backbone with AMP-resistence.
- Team Activity Tests:
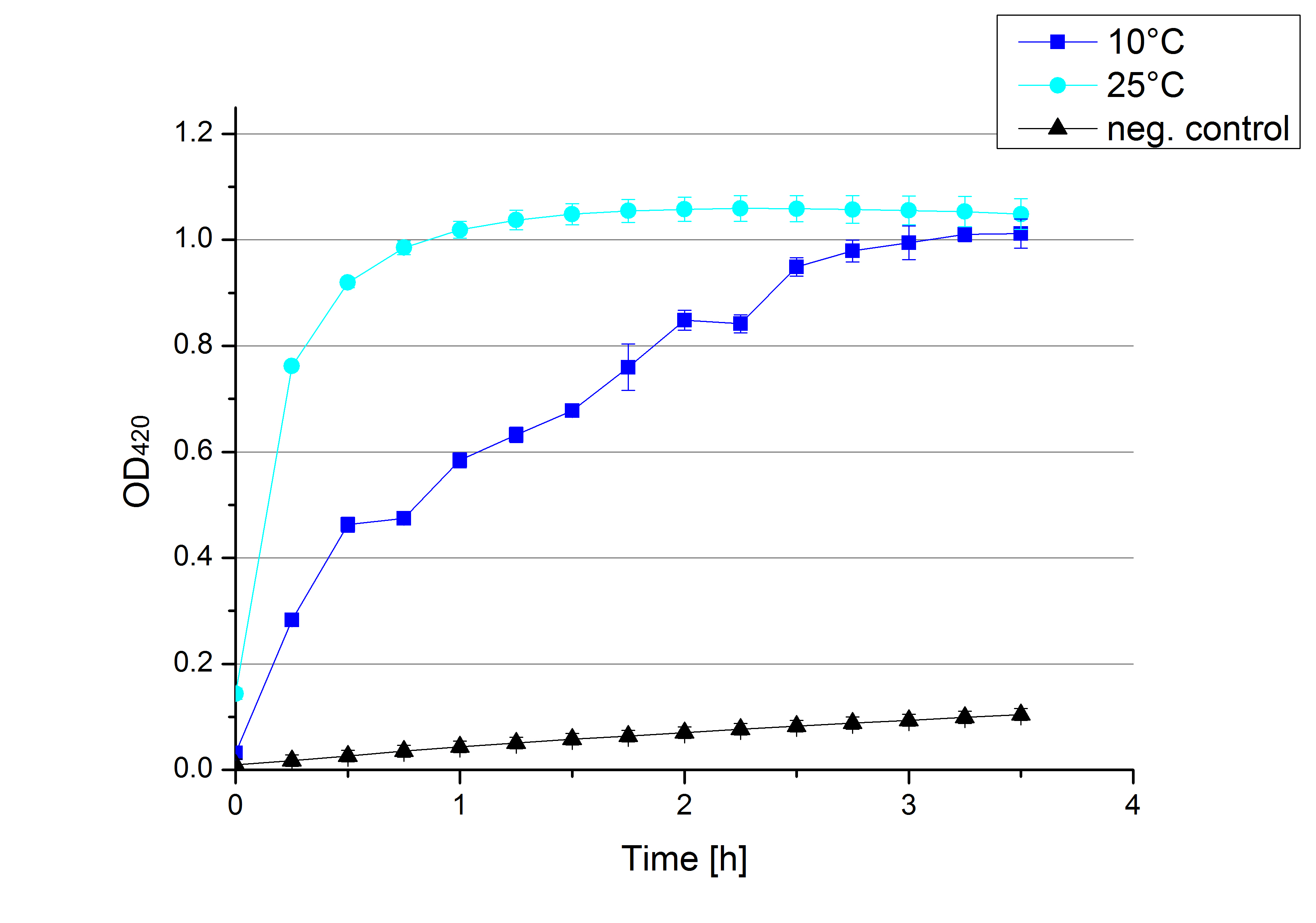
- Today we started another exciting measurement. Today was all about temperatures. Through our cooperation with the clarification plant we found out that the waster water here in Germany has a minimal temperature of 10°C throughout the year. We used this value together with our previous measurements at 25°C to complete the temperature gradient of our measurements. Eventhough [http://partsregistry.org/Part:BBa_K863005 ECOL] shows an increased activity at 10°C, it does reach a comparable OD value as the samples measured at 25°C. The increased activity is expressed by achieving the maximal OD420 slower than the laccase measured at 25°C. This seems like good news for Team Modeling.
- Team Substrate Analytics:
- Degradation measure from Ethin estradiol and Estradiol with the Spectrofluorophotometer. Degradation was from the 18th September
Friday September 21st
- Team Cellulose Binding Domain:
- Transformated <partinfo>J61101</partinfo> with CBDclos+GFP again and plated on the right selection-agar this time!
- Team Activity Tests:
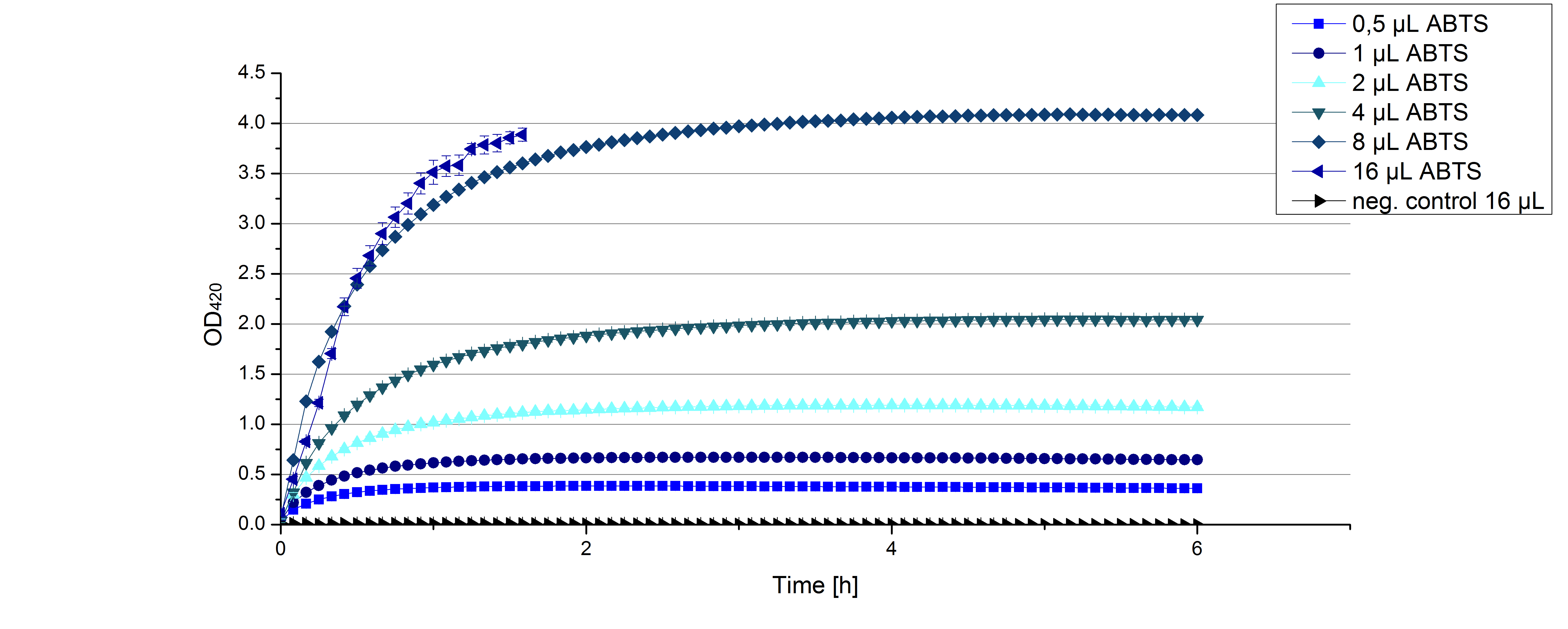
- Another part of the characterization of our [http://partsregistry.org/Part:BBa_K863005 ECOL] and [http://partsregistry.org/wiki/index.php/Part:BBa_K863000 BPUL] laccase was to measure their activity when exposed to different concentrations of ABTS. We choose an exponential gradient of 2 µL to 16 µL and measured with the usual laccase concentration of 0,003 mg/mL and 100 mM sodium acetate buffer (ad 200 µL H2O). Unfortunately the measurements that contained 16 µL ABTS could not be measured completely because the OD420 exceeded the maximal value that Tecan was able to measure. The results of [http://partsregistry.org/Part:BBa_K863005 ECOL] (see. Fig. 1) show as well an exponential increase in the OD420 correlated to the ABTS concentrations, respectively. The negative control was chosen to contain the maximum of 16 µL ABTS in buffer and water that was incubated with CuCl.
Saturday September 22nd
- Team Cellulose Binding Domain:
- Once again two colonies on the agar-dish: can not say if they have a green glow...
- picked the colonies and them put into AMP-LB-Medium.
- Also made a flask with 10 mL LM-Medium and Cells with the [http://partsregistry.org/Part:BBa_K863104 CBDcex(J61101)+GFP_His]-plasmid
- Once again two colonies on the agar-dish: can not say if they have a green glow...
Sunday September 23rd
- Team Cellulose Binding Domain:
- All cultures did not show any sign of GFP-glow under uv-light.
- Gradient-PCR of <partinfo>BBa_K863104</partinfo> with S3N10_Cex_compl- and S3N10_GFP-primermix to generate a 13 amino acid-linker between the cellulose binding domain and the GFP.
- Transformated [http://partsregistry.org/Part:BBa_K863103 CBDcex(T7)+GFP_His] and [http://partsregistry.org/Part:BBa_K863113 CBDclos(T7)+GFP_His] into BL21, to test if the problem is in the expression-system.
| 55px | | | | | | | | | | |
 "
"