Team:Bielefeld-Germany/Protocols/Materials
From 2012.igem.org
Materials
This is where we are going to list all our materials, devices and equipment that we have used.
Devices
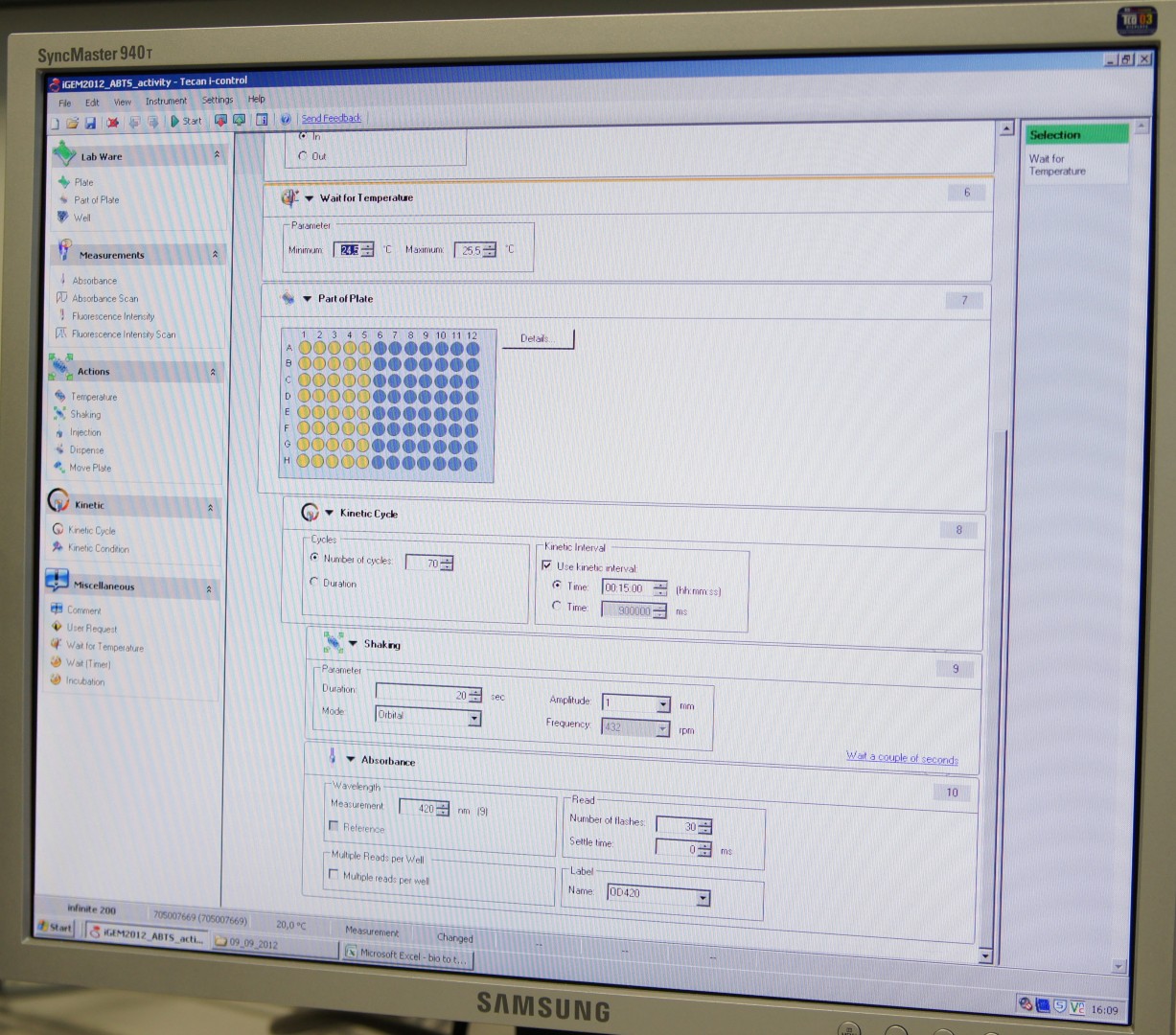
Tecan Infinite Microplate Reader
For measuring the Laccase activity we detected the level of oxidized ABTS via optical density at 420nm. The device we were able to use was a [http://www.tecan.com/platform/apps/product/index.asp?MenuID=1812&ID=1916&Menu=1&Item=21.2.10.1 Tecan Infinite Reader M200]. The program setup was in some parts adapted to the needs of our probes (like duration of the measurement) and in some parts standardized.
Used setup for Laccase activity measurements: Temperature: 25°C; Orbital shaking before each measuring cycle (time depends on duration of each cycle); Number of flashes: 30
Media, buffer and other solutions
Ampicillin stock solution
- Solubilize 100 mg mL-1 Ampicillin
- Store at -20 °C
Chloramphenicol stock solution
- Solubilize 20 mg mL-1 Chloramphenicol in 100 % Ethanol
- Store at -20 °C
TAE buffer
For 1 L of 50 x TAE buffer you need:
- 242.48 g Tris
- 41.02 g Sodiumacetate
- 18.612 g EDTA
- Adjust pH to 7.8 with acetic acid
- Solve in dH2O
10 mL of the stock is diluted in 1 L dH2O for the gel electrophoresis (0.5 x TAE buffer).
Briton Robinson Buffer
- 0,1 mM acetic acid
- 0,1 mM boric acid
- 0,1 mM phosphoric acid
- adjust to pH 5 with sodium hydroxide
DNA loading buffer
- 50 % (v/v) glycerol
- 1 mM EDTA
- 0.1 % (w/v) bromphenol blue
- Solve in ddH2O
LB medium
For 1 L of LB medum:
- 10 g Trypton
- 5 g Yeast extract
- 10 g NaCl
- 12 g Agar-Agar (for plates)
- Adjust pH to 7.4
Autoinduction medium
The Autoinduction medium is based on LB-medium. Add the following components after heat sterilization of 900 mL LB medium.
- 5 mL of a 200 g L-1 steril L-rhamnose stock solution -> final concentration 2 g L-1
- 2.5 mL of a 200 g L-1 steril glucose stock solution -> final concentration 1 g L-1
- if necessary add antibiotics:
- Cm: 1 mL or 3 mL of a 20 μg mL-1 Cm stock solution -> final concentration 20 or 60 mg L-1
- Amp: 1 mL or 3 mL of a 100 μg mL-1 Amp stock solution -> final concentration 100 or 300 mg L-1
- fill up to 1 L with steril ddH2O
YPD media
For 1 L of YPD media:
- 20 g Peptone
- 10 g Yeast extract
- 20 g Dextrose (add 50 mL sterile stock solution (40% dextrose))
- Adjust pH to 6.5
BEDS buffer
For 1 L of BEDS buffer:
- 1,85 g bicine-sodium salt,
- 30 mL ethylene glycol (final concentration 3%(v/v))
- 50 mL (v/v) dimethyl sulfoxide (DMSO)(final concentration 5%(v/v))
- 182.17 g sorbitol
- Adjust pH at 8,3
Dithiothreitol-Solution (DTT)
For 50mL of DTT-Solution:
- 7.71 g Dithiothreitol
Buffers for His-Tag affinity chromatography
- Adjust pH to 7.4 - 7.6
Buffer for Ni-NTA-HisTag affinity chromatography
| Buffer | Sodium phosphate [mM] | Imidazole [mM] |
|---|---|---|
| Binding buffer | 50 | 20 |
| Elution buffer | 50 | 500 |
Buffer for Talon-HisTag affinity chromatography
| Buffer | Sodium phosphate [mM] | NaCl [mM] | Imidazole [mM] |
|---|---|---|---|
| Binding buffer | 50 | 300 | 0 |
| Elution buffer | 50 | 300 | 150 |
SDS-PAGE gel
The following amouts are for one gel. Stacking gel 5 %:
- 775 μL H2O
- 1.25 mL 0,25 M Tris (pH 6,8)
- 425 μL Bis/Acrylamide (0,8 %, 30 %)
- 50 μL 5 % SDS
- 25 μL 10 % Ammonium persulfate
- 3 μL TEMED
Separating gel 12 %:
- 1.5 mL H2O
- 2.8 mL 1 M Tris (pH 8,8)
- 3.0 mL Bis/ Acrylamide (0,8%, 30%)
- 150 μL 5% SDS
- 37.5 μL 10% Ammonium persulfate
- 5 μL TEMED
SDS running buffer
- 25 mM Tris [pH 8,3]
- 192 mM Glycerol
- 0.1 % SDS
4x Laemmli-buffer
- 250 mM Tris-HCl
- 40 % [v/v] Glycerol
- 20 % [v/v] 2-Mercapthoethanol
- 80 g L-1 SDS
- 0.04 g L-1 BPB
Primers
This is a list of primers we have used.
| primer name | length | sequence | |
|---|---|---|---|
| F | 28 | GTTTCTTCGAATTCGCGGCCGCTTCTAG | |
| R | 29 | GTTTCTTCCTGCAGCGGCCGCTACTAGTA | |
| pSB1C3-5aox1-f | 60 | CGCTAAGGATGATTTCTGGAATTCGCGGCCGCTTCTAGAGAGATCTAACATCCAAAGACG | |
| pSB1C3-5aox1-r | 30 | GGTGGCGGCGGGCGTTTCGAATAATTAGTT | |
| 5aox1-mfalpha1-f | 68 | AGAAGATCAAAAAACAACTAATTATTCGAAACGCCCGCCGCCACCATGAGATTTCCTTCAATTTTTAC | |
| 5aox1-mfalpha1-r | 20 | AGCTTCAGCCTCTCTTTTCT | |
| mfalpha1-aarI-taox1-f | 80 | GTATCTCTCGAGAAAAGAGAGGCTGAAGCTACACGCAGGTGGTATGTATCACCTGCGTGTCTTGCTAGAT TCTAATCAAG | |
| mfalpha1-aarI-taox1-r | 20 | TAAGCTTGCACAAACGAACT | |
| taox1-phis4-f | 60 | GTACAGAAGATTAAGTGAGAAGTTCGTTTGTGCAAGCTTATCATGCCATGGACAAGATTC | |
| taox1-phis4-r | 20 | GGCCGCTCGAGTATTCAGAA | |
| phis4-kozak-his4-f | 72 | AATAGTTTACAAAATTTTTTTTCTGAATACTCGAGCGGCCCCCGCCGCCACCATGACATTTCCCTTGCTACC | |
| phis4-kozak-his4-r | 30 | TTATTATTTCTCCATACGAACCTTAACAGC | |
| his4-3aox1-f | 60 | TCACCGCAATGCTGTTAAGGTTCGTATGGAGAAATAATAACGAGTATCTATGATTGGAAG | |
| his4-3aox1-r | 20 | AAAACAAGATAGTGCCCCTC | |
| 3aox1-pSB1C3-f | 60 | AGTCTGATCCTCATCAACTTGAGGGGCACTATCTTGTTTTTACTAGTAGCGGCCGCTGCA | |
| 3aox1-pSB1C3-r | 20 | CTCTAGAAGCGGCCGCGAAT | |
| taox-his4-f | 61 | GTACAGAAGATTAAGTGAGAAGTTCGTTTGTGCAAGCTTAAGATCTCCTGATGACTGACTC | |
| taox-his4-r | 27 | CTCGGATCTATCGAATCTAAATGTAAG | |
| his4-3aox1-f02 | 60 | TTATTTAGAGATTTTAACTTACATTTAGATTCGATAGATCCGAGTATCTATGATTGGAAG | |
| his4_gi537483_f | 46 | ACGTgaattcgcggccgcttctagagAGATCTCCTGATGACTGACT | |
| his4_gi537483_r | 41 | ctgcagcggccgctactagtaGATCTATCGAATCTAAATGT | |
| B.pumi_LAC_FW | 44 | ACGTGAATTCGCGGCCGCTTCTAGATGAACCTAGAAAAATTTGT | |
| B.pumi_LAC_RV | 41 | CTGCAGCGGCCGCTACTAGTATTACTGGATGATATCCATCG | |
| B.halo_FW | 44 | ACGTGAATTCGCGGCCGCTTCTAGATGAAAAAATCATATGGAGT | |
| B.halo_RV | 41 | CTGCAGCGGCCGCTACTAGTATTACTCAGGCATATTTGGAA | |
| T.thermo_LAC_FW | 44 | ACGTGAATTCGCGGCCGCTTCTAGATGCTGGCGCGCAGGAGCTT | |
| T.thermo_LAC_RV | 41 | CTGCAGCGGCCGCTACTAGTACTAACCCACCTCGAGGACTC | |
| E.coli_LAC_FW_T7 | 79 | ACGTGAATTCGCGGCCGCTTCTAGAGtaatacgactcactatagggagagaggagaaaaATGCAACGTCGTGATTTCTT | |
| E.coli_LAC_RV_HIS | 62 | CTGCAGCGGCCGCTACTAGTATTATTAGTGATGGTGATGGTGATGTACCGTAAACCCTAACA | |
| Xcc_LAC_FW_T7 | 79 | ACGTGAATTCGCGGCCGCTTCTAGAGtaatacgactcactatagggagagaggagaaaaATGTCATTCGATCCCTTGTC | |
| Xcc_LAC_RV_HIS | 62 | CTGCAGCGGCCGCTACTAGTATTATTAGTGATGGTGATGGTGATGTGCCTCCACCCGCACTT | |
| B.pumi_LAC_FW_T7 | 79 | ACGTGAATTCGCGGCCGCTTCTAGAGtaatacgactcactatagggagagaggagaaaaATGAACCTAGAAAAATTTGT | |
| B. pumi_LAC_RV_HIS | 62 | CTGCAGCGGCCGCTACTAGTATTATTAGTGATGGTGATGGTGATGCTGGATGATATCCATCG | |
| E.coli_LAC_FW_T7 | 79 | ACGTGAATTCGCGGCCGCTTCTAGAGtaatacgactcactatagggagagaggagaaaaATGCAACGTCGTGATTTCTT | |
| E.coli_LAC_RV_HIS | 62 | CTGCAGCGGCCGCTACTAGTATTATTAGTGATGGTGATGGTGATGTACCGTAAACCCTAACA | |
| T.thermo_LAC_FW_T7 | 79 | ACGTGAATTCGCGGCCGCTTCTAGAGtaatacgactcactatagggagagaggagaaaaATGCTGGCGCGCAGGAGCTT | |
| T.thermo_LAC_RV_HIS | 62 | CTGCAGCGGCCGCTACTAGTATTATTAGTGATGGTGATGGTGATGACCCACCTCGAGGACTC | |
| Pc_lac35_FW_oS | 38 | acgtcacctgcgtgtagctgccatagggcctgtggcgg | |
| Pc_lac35_RV | 39 | acgtcacctgcgtgtcaagTCAGAGGTCGCTGGGGTCAA | |
| Tc_lac5_FW_oS | 38 | acgtcacctgcgtgtagctggtatcggtcctgtcgccg | |
| Tc_lac5_RV | 39 | acgtcacctgcgtgtcaagTTACTGGTCGCTCGGGTCGC | |
| Tv_lac5.P.FW | 43 | gaattcgcggccgcttctagATGTCGAGGTTTCACTCTCTTCT | |
| Tv_lac5.S.RV | 41 | CTGCAGCGGCCGCTACTAGTATTACTGGTCGCTCGGGTCGC | |
| Pc_lac35.P.FW | 43 | gaattcgcggccgcttctagATGTCGAGGTTCCAGTCCCTCTT | |
| Pc_lac35.S.RV | 41 | CTGCAGCGGCCGCTACTAGTATCAGAGGTCGCTGGGGTCAA | |
| J23117_RBS_FW | 70 | aattcgcggccgcttctagagttgacagctagctcagtcctagggattgtgctagcaaagaggagaaata | |
| J23117_RBS_RV | 70 | ctagtatttctcctctttgctagcacaatccctaggactgagctagctgtcaactctagaagcggccgcg | |
| J23103_RBS_FW | 70 | aattcgcggccgcttctagagctgatagctagctcagtcctagggattatgctagcaaagaggagaaata | |
| J23103_RBS_RV | 70 | ctagtatttctcctctttGCTAGCATAATCCCTAGGACTGAGCTAGCTATCAGctctagaagcggccgcg | |
| J23110_RBS_FW | 70 | aattcgcggccgcttctagagtttacggctagctcagtcctaggtacaatgctagcaaagaggagaaata | |
| J23110_RBS_RV | 70 | ctagtatttctcctctttgctagcattgtacctaggactgagctagccgtaaactctagaagcggccgcg | |
| J23103_K_FW | 25 | CTGACAGCTAGCTCAGTCCTAGGTA | |
| J23110/117_K_FW | 25 | TTTACGGCTAGCTCAGTCCTAGGTA | |
| T7_K_FW | 26 | TAATACGACTCACTATAGGGAAAGAG | |
| CBDcex_2AS-Link_Frei | 56 | CTGCAGCGGCCGCTACTAGTATTAACCGGTGCTGCCGCCGACCGTGCAGGGCGTGC | |
| CBDcex_Freiburg-Prefix | 54 | GCTAGAATTCGCGGCCGCTTCTAGATGGCCGGCGGTCCGGCCGGGTGCCAGGTG | |
| CBDcex_T7RBS | 80 | TGAATTCGCGGCCGCTTCTAGAGTAATACGACTCACTATAGGGAAAGAGGAGAAATAATGGGT CCGGCCGGGTGCCAGGT | |
| CBDclos_2ASlink_compl | 63 | CTGCAGCGGCCGCTACTAGTATTAACCGGTGCTGCCTGCAAATCCAAATTCAACATATGTATC | |
| CBDclos_Freiburg-Prefix | 57 | GCTAGAATTCGCGGCCGCTTCTAGATGGCCGGCTCATCAATGTCAGTTGAATTTTAC | |
| Cex_Freiburg_compl | 54 | ACGTCTGCAGCGGCCGCTACTAGTATTAACCGGTGCCGACCGTGCAGGGCGTGC | |
| Clos_Freiburg_compl | 56 | ACGTCTGCAGCGGCCGCTACTAGTATTAACCGGTTGCAAATCCAAATTCAACATAT | |
| Eco_Freiburg | 53 | ACGTGAATTCGCGGCCGCTTCTAGATGGCCGGCCAACGTCGTGATTTCTTAAA | |
| Eco_Freiburg_compl | 53 | ACGTCTGCAGCGGCCGCTACTAGTATTAACCGGTTACCGTAAACCCTAACATC | |
| GFP_Freiburg | 54 | ACGTGAATTCGCGGCCGCTTCTAGATGGCCGGCCGTAAAGGAGAAGAACTTTTC | |
| GFP_Freiburg_compl | 61 | ACGTCTGCAGCGGCCGCTACTAGTATTAACCGGTTTTGTATAGTTCATCCATGCCATGTGT | |
| GFP_His6_compl | 74 | CTGCAGCGGCCGCTACTAGTATTAACCGGTGTGATGGTGATGGTGATGTTTGTATAGTTCATCCATGCCATGTG | |
| GFP_FW | 48 | ATGCGAATTCGCGGCCGCTTCTAGAGTCCCTATCAGTGATAGAGATTG | |
| S3N10_Cex_compl | 40 | TTGTTGTTGTTCGAGCTCGAGCCGACCGTGCAGGGCGTGC | |
| S3N10_Clos_compl | 40 | TTGTTGTTGTTCGAGCTCGAGCTGCCGCCGACCGTGCAGG | |
| S3N10_GFP | 40 | CAATAACAATAACAACAACCGTAAAGGAGAAGAACTTTTC |
Substrates
Hormones
Estradiol Estrone Ethynylestradiol
Analgesics
Diclofenac Ibuprofen Naproxen
Polycyclic aromatic hydrocarbons
Naphthalin Acenaphthen Phenanthren Anthracen
| 55px | | | | | | | | | | |
 "
"